








Biotechnology Bulletin ›› 2022, Vol. 38 ›› Issue (10): 273-280.doi: 10.13560/j.cnki.biotech.bull.1985.2021-1564
Previous Articles Next Articles
WANG Hai-jie( ), WANG Cheng-ji, GUO Yang, WANG Yun, CHEN Yan-juan, LIANG Min, WANG Jue, GONG Hui, SHEN Ru-ling(
), WANG Cheng-ji, GUO Yang, WANG Yun, CHEN Yan-juan, LIANG Min, WANG Jue, GONG Hui, SHEN Ru-ling( )
)
Received:2022-02-21
Online:2022-10-26
Published:2022-11-11
Contact:
SHEN Ru-ling
E-mail:wanghaijie@slarc.org.cn;shenruling@slarc.org.cn
WANG Hai-jie, WANG Cheng-ji, GUO Yang, WANG Yun, CHEN Yan-juan, LIANG Min, WANG Jue, GONG Hui, SHEN Ru-ling. Construction of Coagulation Factor 8 Gene Knockout Mouse Model Based on CRSIPR/Cas9 Technique and Verification of Phenotype[J]. Biotechnology Bulletin, 2022, 38(10): 273-280.
| sgRNA靶序列 sgRNA target sequence | 序列信息 Sequence information(5'-3') |
|---|---|
| sgRNA 1 | AGGCTTAACCCATTTCCTGC TGG |
| sgRNA 2 | TCTGGATCCACAGATATAGC AGG |
Table 1 sgRNA sequence information
| sgRNA靶序列 sgRNA target sequence | 序列信息 Sequence information(5'-3') |
|---|---|
| sgRNA 1 | AGGCTTAACCCATTTCCTGC TGG |
| sgRNA 2 | TCTGGATCCACAGATATAGC AGG |
| 寡核苷酸OligoDNA | 序列信息Sequence information(5'-3') |
|---|---|
| sgRNA 1 正义链 | ACTTATTAGTTTGCAAAACG |
| sgRNA 1 反义链 | GTGGCCTTCGATTAGGCGGA |
| sgRNA 2 正义链 | CGGTCAGAGCTGCACATACA |
| sgRNA 2 反义链 | GCCAAATTGTGATCTCAGTT |
Table 2 Oligonucleotide chain sequence information
| 寡核苷酸OligoDNA | 序列信息Sequence information(5'-3') |
|---|---|
| sgRNA 1 正义链 | ACTTATTAGTTTGCAAAACG |
| sgRNA 1 反义链 | GTGGCCTTCGATTAGGCGGA |
| sgRNA 2 正义链 | CGGTCAGAGCTGCACATACA |
| sgRNA 2 反义链 | GCCAAATTGTGATCTCAGTT |
| 引物Primer | 序列信息Sequence information(5'-3') |
|---|---|
| P1 | TTTTGGCTTTCTAACAGGTATCG |
| P2 | CCAGAAAAGGGCATCAGGT |
Table 3 Primer sequence information
| 引物Primer | 序列信息Sequence information(5'-3') |
|---|---|
| P1 | TTTTGGCTTTCTAACAGGTATCG |
| P2 | CCAGAAAAGGGCATCAGGT |
| 基因 Gene | 上游引物信息Upstream primer information(5'-3') | 下游引物信息Downstream primer information(5'-3') |
|---|---|---|
| F8 | AGGCCTCATTGGAGCTCTGCTA | CCATTGGGATTCCTCAGGGCA |
| β-actin | CGTTGACATCCGTAAAGACC | AACAGTCCGCCTAGAAGCAC |
Table 4 Reverse transcription PCR primer sequences
| 基因 Gene | 上游引物信息Upstream primer information(5'-3') | 下游引物信息Downstream primer information(5'-3') |
|---|---|---|
| F8 | AGGCCTCATTGGAGCTCTGCTA | CCATTGGGATTCCTCAGGGCA |
| β-actin | CGTTGACATCCGTAAAGACC | AACAGTCCGCCTAGAAGCAC |
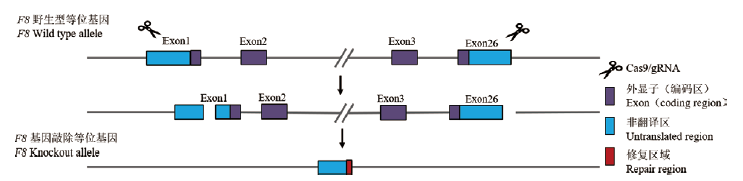
Fig.1 Knockout strategy of F8 -/- mice The Cas9/sgRNA complex removed the corresponding fragments of the F8 gene by recognizing targets in the exon1-26 region
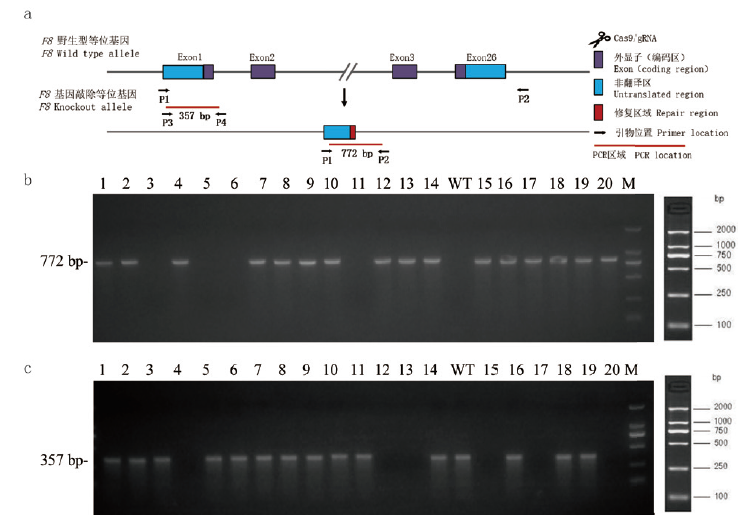
Fig.2 Genotype results of F8-/- gene knockout mice (a)The identification strategy of F8-/- knockout mice. Two pairs of primers P1/P2 and P3/P4 were used for PCR identification to determine the mice genotype. The primer positions were shown in the figure.(b)and(c)were genotype identification electrophoresis diagram,with P1/P2 primer pair for PCR detection in(b),bands were obtained in WT mice,F8-/+ heterozygous mice and F8-/- homozygous mice obtained a single 772 bp fragment.(c)The P3/P4 primer pair was used for the PCR detection,WT mice and F8-/+ heterozygous mice could obtain a 357 bp fragment,F8-/- homozygous mice could not obtain a band. According to electrophoresis identification,WT mice in(b)and(c)were numbered 3,5,6,11,and F8-/+ heterozygous mice were numbered 1,2,7,8,9,10,14,16,18,19,F8-/- homozygous mice were numbered 4,12,13,15,17,20. WT:Wild type mice. M:1 kb DNA maker
| 类型Index | 序列信息Sequence information(5'-3') |
|---|---|
| 野生型小鼠 WT mice | AGGATTCAAACTTGTTAGGATGCACCCAGCAGGA- AATGGGTTAAGCCTTAGCTCAGCCACTCTTCCTA- TTCCAGTT……GAGAAGTTGCTGAGAGTTCTATA- TCTGGATCCACAGATATAGCAGGAAGAGAAAGA- CACTGGGACTGACTTGGG |
| F8-/-小鼠序列 F8-/-mice | AGGATTCAAACTTGTTAGGATGCACCCAGCAGGAA …(207293 bp缺失)…GAGAAAGACACTGGGAC- TGACTTGGG |
Table 5 Sequence information of wild type and F8-/-mice genotype
| 类型Index | 序列信息Sequence information(5'-3') |
|---|---|
| 野生型小鼠 WT mice | AGGATTCAAACTTGTTAGGATGCACCCAGCAGGA- AATGGGTTAAGCCTTAGCTCAGCCACTCTTCCTA- TTCCAGTT……GAGAAGTTGCTGAGAGTTCTATA- TCTGGATCCACAGATATAGCAGGAAGAGAAAGA- CACTGGGACTGACTTGGG |
| F8-/-小鼠序列 F8-/-mice | AGGATTCAAACTTGTTAGGATGCACCCAGCAGGAA …(207293 bp缺失)…GAGAAAGACACTGGGAC- TGACTTGGG |
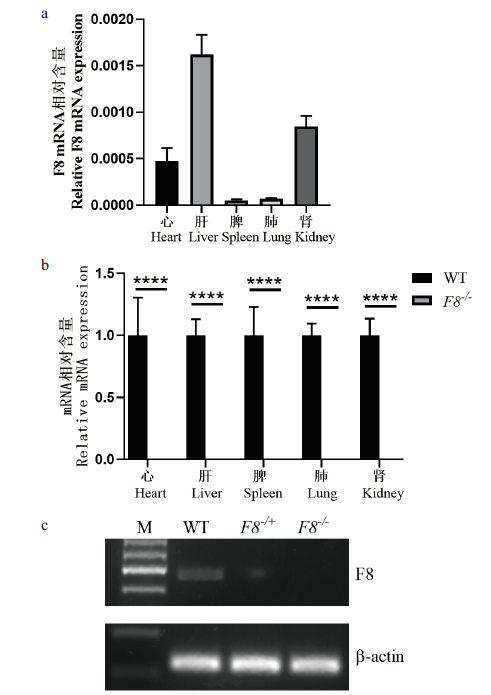
Fig.3 Analysis of relative mRNA expression in F8-/- mice (a)F8 gene expression in multiple tissues of WT mice.(b)F8 mRNA relative expression level detection in heart,liver,spleen,lung and kidney tissues of F8 -/- and WT mice,3 mice per group(****P <0.0001).(c)RT-PCR representative identification result of electrophoresis(liver tissue). WT:Wild type mice,F8 -/+-heterozygous,F8 -/--homozygous mice
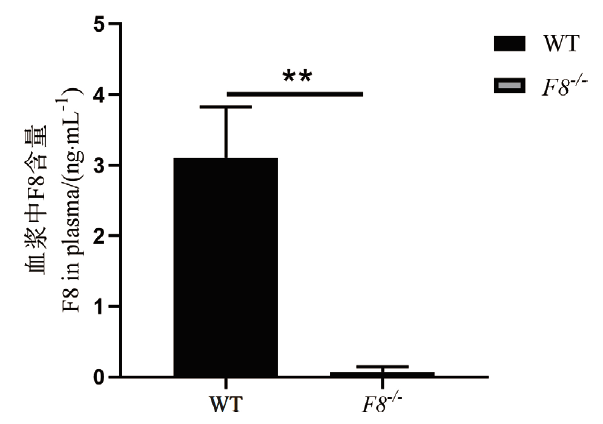
Fig.4 Relative expression of F8 protein in F8-/- mice F8 -/- protein content in plasma of F8-/- and WT mice,5 mice per group(**P <0.01),WT-wild type,F8-/-- homozygous
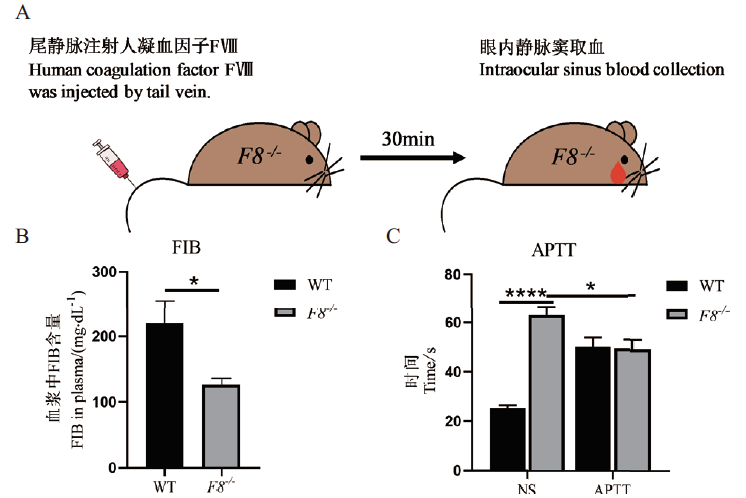
Fig.5 APTT results of F8-/- mice A:Schematic diagram of the administration process of blood coagulation in mice. B:Comparison of FIB in plasma of F8 -/- and WT mice. C:Comparison of APTT values before and after administration of human coagulation factor Ⅷ between F8 -/- and WT mice,5 mice per group(*P <0.05,****P <0.0001),to NS-tail vein injected with saline,to APTT-tail vein injected with human coagulation factor Ⅷ,WT-wild type,and F8 -/-- homozygous
| [1] |
Jankowska KI, Chattopadhyay M, Sauna ZE, et al. A foundational study for normal F8-containing mouse models for the miRNA regulation of hemophilia A:identification and analysis of mouse miRNAs that downregulate the murine F8 gene[J]. Int J Mol Sci, 2020, 21(16):5621.
doi: 10.3390/ijms21165621 URL |
| [2] | 王典文, 涂传清. 血友病A发病分子机制的研究现状[J]. 医学综述, 2013, 19(24):4430-4433. |
| Wang DW, Tu CQ. Research status of molecular pathogenesis of hemophilia A[J]. Med Recapitul, 2013, 19(24):4430-4433. | |
| [3] | 许正平, 王恩多. 凝血因子Ⅷ[J]. 生物化学与生物物理进展, 1991(4):8-11. |
| Xu ZP, Wang ED. Coagulation factor Ⅷ[J]. Prog Biochem Biophys, 1991(4):8-11. | |
| [4] | 侯玉香, 肖小璞. 血友病基因治疗概述[J]. 中国输血杂志, 2007, 20(1):71-74. |
| Hou YX, Xiao XP. Summary of gene therapy for hemophilia[J]. Chin J Blood Transfus, 2007, 20(1):71-74. | |
| [5] |
Yamaguti-Hayakawa GG, Ozelo MC. Gene therapy:paving new roads in the treatment of hemophilia[J]. Semin Thromb Hemost, 2019, 45(7):743-750.
doi: 10.1055/s-0039-1688445 pmid: 31096314 |
| [6] |
Rangarajan S, Walsh L, Lester W, et al. AAV5-factor VIII gene transfer in severe hemophilia A[J]. N Engl J Med, 2017, 377(26):2519-2530.
doi: 10.1056/NEJMoa1708483 URL |
| [7] |
Batty P, Lillicrap D. Advances and challenges for hemophilia gene therapy[J]. Hum Mol Genet, 2019, 28(R1):R95-R101.
doi: 10.1093/hmg/ddz157 |
| [8] |
Monahan PE, Samulski RJ, Tazelaar J, et al. Direct intramuscular injection with recombinant AAV vectors results in sustained expression in a dog model of hemophilia[J]. Gene Ther, 1998, 5(1):40-49.
pmid: 9536263 |
| [9] |
Peyvandi F, Garagiola I. Clinical advances in gene therapy updates on clinical trials of gene therapy in haemophilia[J]. Haemophilia, 2019, 25(5):738-746.
doi: 10.1111/hae.13816 pmid: 31282050 |
| [10] |
Chen HN, Shi M, Gilam A, et al. Hemophilia a ameliorated in mice by CRISPR-based in vivo genome editing of human factor VIII[J]. Sci Rep, 2019, 9(1):16838.
doi: 10.1038/s41598-019-53198-y URL |
| [11] |
Guo T, Feng YL, Xiao JJ, et al. Harnessing accurate non-homologous end joining for efficient precise deletion in CRISPR/Cas9-mediated genome editing[J]. Genome Biol, 2018, 19(1):170.
doi: 10.1186/s13059-018-1518-x pmid: 30340517 |
| [12] | Hooper ML. Embryonic stem cells:introducing planned changes into the animal germline[M]. Chur: Harwood Academic Publishers, 1992. |
| [13] |
Lin HF, Maeda N, Smithies O, et al. A coagulation factor IX-deficient mouse model for human hemophilia B[J]. Blood, 1997, 90(10):3962-3966.
pmid: 9354664 |
| [14] | 陈永昌, 牛昱宇, 季维智. 通过CRISPR/Cas9和TALENs介导的基因打靶技术获得基因修饰的猴模型[J]. 中国细胞生物学学报, 2014, 36(5):557-560. |
| Chen YC, Niu YY, Ji WZ. The genetically modified monkey model was obtained by gene targeting mediated by CRISPR/ Cas9 and TALENs[J]. Chin J Cell Biol, 2014, 36(5):557-560. | |
| [15] |
Cong L, Ran FA, Cox D, et al. Multiplex genome engineering using CRISPR/Cas systems[J]. Science, 2013, 339(6121):819-823.
doi: 10.1126/science.1231143 pmid: 23287718 |
| [16] | 梁振伟, 饶书权, 沈岩, 等. 通过CRISPR/Cas9系统敲除人源PDE10A基因[J]. 基础医学与临床, 2014, 34(4):439-443. |
| Liang ZW, Rao SQ, Shen Y, et al. Knocking out human PDE10A gene by CRISPR/Cas9 system[J]. Basic Clin Med, 2014, 34(4):439-443. | |
| [17] | 郑武, 谷峰. CRISPR/Cas9的应用及脱靶效应研究进展[J]. 遗传, 2015, 37(10):1003-1010. |
| Zheng W, Gu F. Progress of application and off-target effects of CRISPR/Cas9[J]. Hereditas, 2015, 37(10):1003-1010. | |
| [18] |
Ma YW, Chen W, Zhang X, et al. Increasing the efficiency of CRISPR/Cas9-mediated precise genome editing in rats by inhibiting NHEJ and using Cas9 protein[J]. RNA Biol, 2016, 13(7):605-612.
doi: 10.1080/15476286.2016.1185591 pmid: 27163284 |
| [19] | Du J, Yin NR, Xie T, et al. Quantitative assessment of HR and NHEJ activities via CRISPR/Cas9-induced oligodeoxynucleotide-mediated DSB repair[J]. DNA Repair(Amst), 2018, 70:67-71. |
| [20] | 汪启翰, 怀聪, 孙瑞林, 等. 利用CRISPR/Cas系统快速高效构建血友病乙小鼠模型[J]. 遗传, 2015, 37(11):1143-1148. |
| Wang QH, Huai C, Sun RL, et al. A quick and efficient method to generate hemophilia B mouse models by the CRISPR/Cas system[J]. Hereditas, 2015, 37(11):1143-1148. |
| [1] | CHEN Xiao-ling, LIAO Dong-qing, HUANG Shang-fei, CHEN Ying, LU Zhi-long, CHEN Dong. Advances in CRISPR/Cas9 System Modifying Saccharomycescerevisiae [J]. Biotechnology Bulletin, 2023, 39(8): 148-158. |
| [2] | YANG Yu-mei, ZHANG Kun-xiao. Establishing a Stable Cell Line with Site-specific Integration of ERK Kinase Phase-separated Fluorescent Probe Using CRISPR/Cas9 Technology [J]. Biotechnology Bulletin, 2023, 39(8): 159-164. |
| [3] | SHI Wei-tao, YAO Chun-peng, WEI Wen-Kang, WANG Lei, FANG Yuan-jie, TONG Yu-jie, MA Xiao-jiao, JIANG Wen, ZHANG Xiao-ai, SHAO Wei. Establishment of MDH2 Knockout Cell Line Using CRISPR/Cas9 Technology and Study of Anti-deoxynivalenol Effect [J]. Biotechnology Bulletin, 2023, 39(7): 307-315. |
| [4] | LIU Xiao-yan, ZHU Zhen-liang, SHI Guang-yu, HUA Zi-yu, YANG Chen, ZHANG Yong, LIU Jun. Strategies to Optimize the Expression of Mammary Gland Bioreactor [J]. Biotechnology Bulletin, 2023, 39(5): 77-91. |
| [5] | CHENG Jing-wen, CAO Lei, ZHANG Yan-min, YE Qian, CHEN Min, TAN Wen-song, ZHAO Liang. Establishment and Application of Multigene Engineering Transformation Strategy for CHO Cells [J]. Biotechnology Bulletin, 2023, 39(2): 283-291. |
| [6] | HUANG Wen-li, LI Xiang-xiang, ZHOU Wen-ting, LUO Sha, YAO Wei-jia, MA Jie, ZHANG Fen, SHEN Yu-sen, GU Hong-hui, WANG Jian-sheng, SUN Bo. Targeted Editing of BoZDS in Broccoli by CRISPR/Cas9 Technology [J]. Biotechnology Bulletin, 2023, 39(2): 80-87. |
| [7] | WANG Bing, ZHAO Hui-na, YU Jing, CHEN Jie, LUO Mei, LEI Bo. Regulation of Leaf Bud by REVOLUTA in Tobacco Based on CRISPR/Cas9 System [J]. Biotechnology Bulletin, 2023, 39(10): 197-208. |
| [8] | LI Shuang-xi, HUA Jin-lian. Research Progress in Anti-porcine Reproductive and Respiratory Syndrome Genetically Modified Pigs [J]. Biotechnology Bulletin, 2023, 39(10): 50-57. |
| [9] | LIN Rong, ZHENG Yue-ping, XU Xue-zhen, LI Dan-dan, ZHENG Zhi-fu. Functional Analysis of ACOL8 Gene in the Ethylene Synthesis and Response in Arabidopsis thaliana [J]. Biotechnology Bulletin, 2023, 39(1): 157-165. |
| [10] | LIU Jing-jing, LIU Xiao-rui, LI Lin, WANG Ying, YANG Hai-yuan, DAI Yi-fan. Establishment of Porcine Fetal Fibroblasts with OXTR-knockout Using CRISPR/Cas9 [J]. Biotechnology Bulletin, 2022, 38(6): 272-278. |
| [11] | Olalekan Amoo, HU Li-min, ZHAI Yun-gu, FAN Chu-chuan, ZHOU Yong-ming. Regulation of Shoot Branching by BRANCHED1 in Brassica napus Based on Gene Editing Technology [J]. Biotechnology Bulletin, 2022, 38(4): 97-105. |
| [12] | DING Ya-qun, DING Ning, XIE Shen-min, HUANG Meng-na, ZHANG Yu, ZHANG Qin, JIANG Li. Construction of Vps28 Knock-out Mice and Model Study of the Impact on Lactation and Immune Traits [J]. Biotechnology Bulletin, 2022, 38(3): 164-172. |
| [13] | YAN Jiong, FENG Chen-yi, GAO Xue-kun, XU Xiang, YANG Jia-min, CHEN Zhao-yang. Construction of Homozygous Plin1-knockout Mouse Model and Phenotype Analysis Based on CRISPR/Cas9 Technology [J]. Biotechnology Bulletin, 2022, 38(3): 173-180. |
| [14] | ZHONG Jing, SUN Ling-ling, ZHANG Shu, MENG Yuan, ZHI Yi-fei, TU Li-qing, XU Tian-peng, PU Li-ping, LU Yang-qing. Effect of Knocking Out the Mda5 Gene by CRISPR/Cas9 Technology on the Replication of Newcastle Disease and Infectious Bursal Virus [J]. Biotechnology Bulletin, 2022, 38(11): 90-96. |
| [15] | ZONG Mei, HAN Shuo, GUO Ning, DUAN Meng-meng, LIU Fan, WANG Gui-xiang. Production of Marker-free Mutants of Brassica campestris Mediated by CRISPR/Cas9 Through Vacuum Infiltration [J]. Biotechnology Bulletin, 2022, 38(10): 159-163. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||