








Biotechnology Bulletin ›› 2022, Vol. 38 ›› Issue (10): 262-272.doi: 10.13560/j.cnki.biotech.bull.1985.2021-1556
Previous Articles Next Articles
ZHENG Qing-bo1,2( ), YE Na1,2, ZHANG Xiao-lan2, BAO Peng-jia2, WANG Fu-bin1,2, REN Wen-wen1,2, LIAO Yue-jiao1, YAN Ping2, PAN He-ping1(
), YE Na1,2, ZHANG Xiao-lan2, BAO Peng-jia2, WANG Fu-bin1,2, REN Wen-wen1,2, LIAO Yue-jiao1, YAN Ping2, PAN He-ping1( )
)
Received:2021-12-16
Online:2022-10-26
Published:2022-11-11
Contact:
PAN He-ping
E-mail:1404871953@qq.com;panheping62@163.com
ZHENG Qing-bo, YE Na, ZHANG Xiao-lan, BAO Peng-jia, WANG Fu-bin, REN Wen-wen, LIAO Yue-jiao, YAN Ping, PAN He-ping. Identification of Hair Follicle Cell Subsets and Bioinformatics Analysis of Characteristic Genes in Tianzhu White Yak During Catagen[J]. Biotechnology Bulletin, 2022, 38(10): 262-272.
| 亚群Cluster ID | 标记基因 Marker gene | 类型Cell type | 参考文献Reference |
|---|---|---|---|
| 0,3,8,9 | KRT14,KRT15,KRT5 | 表皮细胞Epidermal | [ |
| 1,4,10 | KRT1,KRT10,KRTDAP,SBSN | 滤泡间上皮分化细胞 Interfollicular epidermis differentiated cell | [ |
| 2,6 | SPARC,LHX2 | 毛干细胞 Hair shaft | [ |
| 5 | KRT28,KRT27,KRT71 | 内根鞘细胞 Inner root sheath | [ |
| 7 | TOP2A,UBE2C | 漏斗细胞 Infundibulum | [ |
| 13 | PMEL,TYRP1 | 黑素细胞Melanocyte | [ |
Table 1 Cell type marker molecules
| 亚群Cluster ID | 标记基因 Marker gene | 类型Cell type | 参考文献Reference |
|---|---|---|---|
| 0,3,8,9 | KRT14,KRT15,KRT5 | 表皮细胞Epidermal | [ |
| 1,4,10 | KRT1,KRT10,KRTDAP,SBSN | 滤泡间上皮分化细胞 Interfollicular epidermis differentiated cell | [ |
| 2,6 | SPARC,LHX2 | 毛干细胞 Hair shaft | [ |
| 5 | KRT28,KRT27,KRT71 | 内根鞘细胞 Inner root sheath | [ |
| 7 | TOP2A,UBE2C | 漏斗细胞 Infundibulum | [ |
| 13 | PMEL,TYRP1 | 黑素细胞Melanocyte | [ |
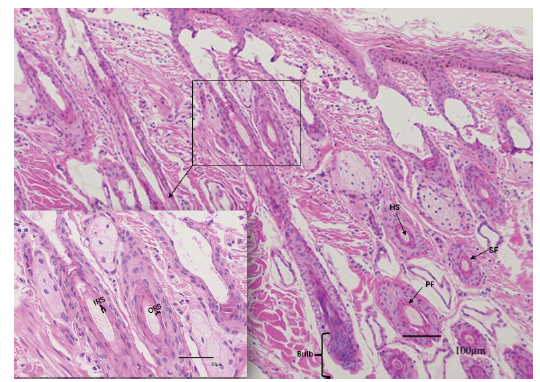
Fig. 1 Skin tissue structure of Tianzhu white yak ORS:Outer root sheath. IRS:Inner root sheath. PF:Primary follicles. SF:Secondary follicles. HS:Hair shaft
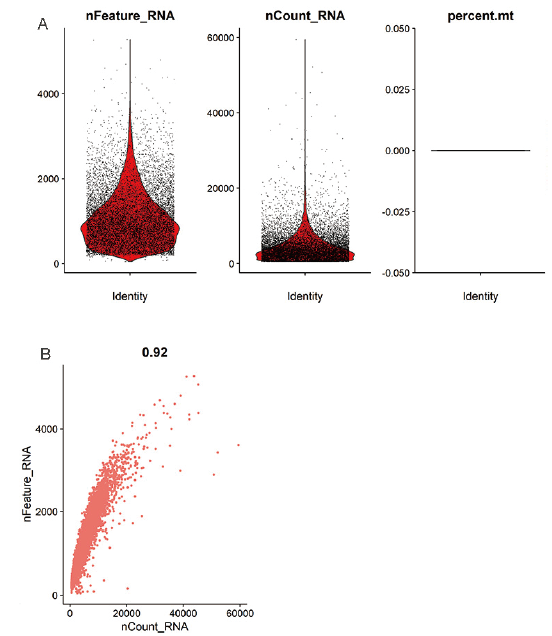
Fig. 2 Statistics and correlation analysis of gene number and total UMI in detected cells A:Number of cell genes and total UMI statistics. B:Correlation analysis between gene number and total UMI. Each point represents a cell,the nFeature _ RNA ordinate represents the number of genes expressed in each cell,nCount _ RNA ordinate represents the total UMI number in each cell,percent.mt ordinate represents the ratio of mitochondrial gene UMI to total UMI in each cell,and the abscissa indicates the abundance of cells in each ordinate value
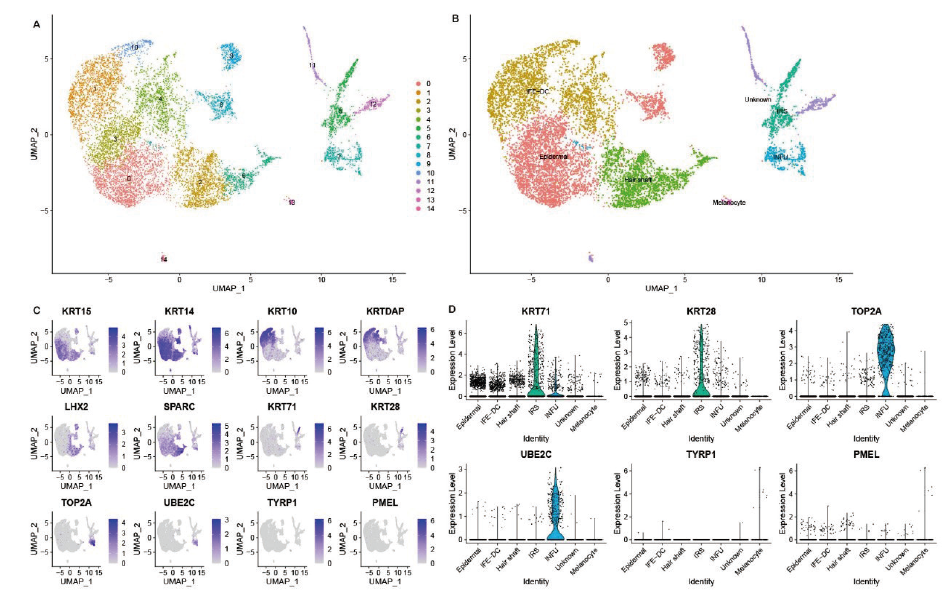
Fig.5 UMAP clustering and identification of major cell types A:Cell clustering information in the UMAP plot. B:Identification of major cell types in UMAP plot. C:The expressions of specific marker genes in different cells. D:Expression of specific markers in some clusters

Fig. 7 Enrichment analysis of different clusters characteristic genes in epidermal cell lineage and IFE-DC A:Enrichment analysis of different clusters characteristic genes in epidermal cell lineage. B:Enrichment analysis of different clusters characteristic genes in IFE-DC

Fig.8 Enrichment analysis of HS and melanocytes characteristic genes A:Enrichment analysis of HS characteristic genes. B:Enrichment analysis of melanocyte characteristic genes
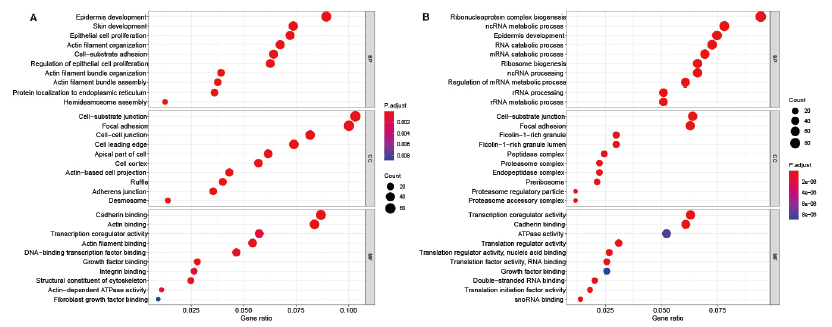
Fig.9 GO enrichment analysis of characteristic genes in INFU and IRS cells A:GO enrichment analysis of INFU characteristic genes. B:GO enrichment analysis of IRS characteristic genes

Fig. 10 KEGG Enrichment analysis of characteristic gene in different cell clusters A:KEGG enrichment map of different cells. B:Characteristic gene interaction network diagram
| [1] | 郭华. 中西医结合对牦牛犊牛腹泻病的诊疗[J]. 中兽医学杂志, 2016(3):49-50. |
| Guo H. Diagnosis and treatment of yak calf diarrhea by integrated traditional Chinese and western medicine[J]. Chin J Tradit Vet Sci, 2016(3):49-50. | |
| [2] | 董巧霞, 石斌刚, 左志, 等. 天祝白牦牛绒毛品质分析[J]. 中国畜牧杂志, 2021, 57(12):110-114. |
| Dong QX, Shi BG, Zuo Z, et al. Cashmere quality analysis of Tianzhu white yak[J]. Chin J Animal Sci, 2021, 57(12):110-114. | |
| [3] |
Han XP, Wang RY, Zhou YC, et al. Mapping the mouse cell atlas by microwell-seq[J]. Cell, 2018, 172(5):1091-1107. e17.
doi: S0092-8674(18)30116-8 pmid: 29474909 |
| [4] |
Tang FC, et al. mRNA-Seq whole-transcriptome analysis of a single cell[J]. Nat Methods, 2009, 6(5):377-382.
doi: 10.1038/nmeth.1315 pmid: 19349980 |
| [5] |
He WY, Ye JG, Xu HY, et al. Differential expression of α6 and β1 integrins reveals epidermal heterogeneity at single-cell resolution[J]. J Cell Biochem, 2020, 121(3):2664-2676.
doi: 10.1002/jcb.29487 pmid: 31680320 |
| [6] |
Yang H, et al. Epithelial-mesenchymal micro-niches govern stem cell lineage choices[J]. Cell, 2017, 169(3):483-496. e13.
doi: S0092-8674(17)30367-7 pmid: 28413068 |
| [7] |
Joost S, et al. The molecular anatomy of mouse skin during hair growth and rest[J]. Cell Stem Cell, 2020, 26(3):441-457. e7.
doi: S1934-5909(20)30012-6 pmid: 32109378 |
| [8] |
Colombo S, et al. Transcriptomic analysis of mouse embryonic skin cells reveals previously unreported genes expressed in melanoblasts[J]. J Invest Dermatol, 2012, 132(1):170-178.
doi: 10.1038/jid.2011.252 pmid: 21850021 |
| [9] | 胡帅帅, 张珉, 肖叶懿, 等. 不同毛色獭兔皮肤中黑色素沉积及毛囊发育规律[J]. 西北农业学报, 2019, 28(9):1387-1393. |
| Hu SS, Zhang M, Xiao YY, et al. Melanin deposition and hair follicle development in skins of rex rabbit with different coat colors[J]. Acta Agric Boreali Occidentalis Sin, 2019, 28(9):1387-1393. | |
| [10] |
Hardy MH. The secret life of the hair follicle[J]. Trends Genet, 1992, 8(2):55-61.
pmid: 1566372 |
| [11] | 谷聪敏. 胎鼠真皮细胞诱导毛发形成的研究[D]. 北京: 北京协和医学院, 2013. |
| Gu CM. Study on hair formation induced by fetal rat dermal cells[D]. Beijing: Peking Union Medical College, 2013. | |
| [12] |
Lindner G, Botchkarev VA, Botchkareva NV, et al. Analysis of apoptosis during hair follicle regression(catagen)[J]. Am J Pathol, 1997, 151(6):1601-1617.
pmid: 9403711 |
| [13] | 韩勇. 不同培养基、药物对体外人游离毛囊生长的影响[D]. 扬州: 扬州大学, 2007. |
| Han Y. The study of cultured human hair follicle growth about different cultured conditions and medicines[D]. Yangzhou: Yangzhou University, 2007. | |
| [14] | 赵桂儒. 较大规模数据应用PCA降维的一种方法[J]. 电脑知识与技术, 2014(8):1835-1837. |
| Zhao GR. A method of dimensionality reduction for large scale data using PCA[J]. Comput Knowl Technol, 2014(8):1835-1837. | |
| [15] | Hwang B, Lee JH, Bang D. Single-cell RNA sequencing technologies and bioinformatics pipelines[J]. Exp Mol Med, 2018, 50(8):1-14. |
| [16] | Maaten LVD, Hinton G. Viualizing data using t-SNE[J]. J Mach Learn Res, 2008, 9(2605):2579-2605. |
| [17] |
Satpathy AT, et al. Massively parallel single-cell chromatin landscapes of human immune cell development and intratumoral T cell exhaustion[J]. Nat Biotechnol, 2019, 37(8):925-936.
doi: 10.1038/s41587-019-0206-z pmid: 31375813 |
| [18] |
Ryan KR, Lock FE, Heath JK, et al. Plakoglobin-dependent regulation of keratinocyte apoptosis by Rnd3[J]. J Cell Sci, 2012, 125(Pt 13):3202-3209.
doi: 10.1242/jcs.101931 pmid: 22454524 |
| [19] |
Fuchs E. Scratching the surface of skin development[J]. Nature, 2007, 445(7130):834-842.
doi: 10.1038/nature05659 URL |
| [20] |
Müller EJ, Williamson L, Kolly C, et al. Outside-in signaling through integrins and cadherins:a central mechanism to control epidermal growth and differentiation?[J]. J Invest Dermatol, 2008, 128(3):501-516.
doi: 10.1038/sj.jid.5701248 URL |
| [21] | 葛伟. 单细胞分辨率解析绒山羊及小鼠毛囊发生的转录调控机制[D]. 杨凌: 西北农林科技大学, 2019. |
| Ge W. Dissecting the transcriptional regulatory mechanism underlying cashmere goat and murine hair follicle morphogenesis at single-cell resolution[D]. Yangling: Northwest A & F University, 2019. | |
| [22] | 叶娜. 基于单细胞转录组测序对天祝白牦牛生长期毛囊转录图谱的构建[D]. 兰州: 西北民族大学, 2021. |
| Ye N. Construction of transcription map of hair follicles in growing period of Tianzhu white yak based on single cell transcriptome sequencing[D]. Lanzhou: Northwest University for Nationalities, 2021. | |
| [23] |
Badamchian M, et al. Thymosin β(4)reduces lethality and down-regulates inflammatory mediators in endotoxin-induced septic shock[J]. Int Immunopharmacol, 2003, 3(8):1225-1233.
doi: 10.1016/S1567-5769(03)00024-9 pmid: 12860178 |
| [24] |
Slominski A, Tobin DJ, Shibahara S, et al. Melanin pigmentation in mammalian skin and its hormonal regulation[J]. Physiol Rev, 2004, 84(4):1155-1228.
pmid: 15383650 |
| [25] | 高泽成. 牦牛毛色候选基因的筛选及MC1R基因功能验证[D]. 兰州: 甘肃农业大学, 2017. |
| Gao ZC. The selection about yak hair color candidate genes and MC1R gene functional verification[D]. Lanzhou: Gansu Agricultural University, 2017. | |
| [26] |
Botchkareva NV, Botchkarev VA, Gilchrest BA. Fate of melanocytes during development of the hair follicle pigmentary unit[J]. J Investig Dermatol Symp Proc, 2003, 8(1):76-79.
doi: 10.1046/j.1523-1747.2003.12176.x URL |
| [1] | WU Hao, LIU Zi-wei, ZHENG Ying, DAI Ya-wen, SHI Quan. Study on the Heterogeneity of Human Gingival Mesenchymal Stem Cells at Single Cell Level [J]. Biotechnology Bulletin, 2023, 39(7): 325-332. |
| [2] | WANG Yi-qing, WANG Tao, WEI Chao-ling, DAI Hao-min, CAO Shi-xian, SUN Wei-jiang, ZENG Wen. Identification and Interaction Analysis of SMAS Gene Family in Tea Plant(Camellia sinensis) [J]. Biotechnology Bulletin, 2023, 39(4): 246-258. |
| [3] | PING Huai-lei, GUO Xue, YU Xiao, SONG Jing, DU Chun, WANG Juan, ZHANG Huai-bi. Cloning and Expression of PdANS in Paeonia delavayi and Correlation with Anthocyanin Content [J]. Biotechnology Bulletin, 2023, 39(3): 206-217. |
| [4] | GUO Zhi-hao, JIN Ze-xin, LIU Qi, GAO Li. Bioinformatics Analysis, Subcellular Localization and Toxicity Verification of Effector g11335 in Tilletia contraversa Kühn [J]. Biotechnology Bulletin, 2022, 38(8): 110-117. |
| [5] | YU Qiu-lin, MA Jing-yi, ZHAO Pan, SUN Peng-fang, HE Yu-mei, LIU Shi-biao, GUO Hui-hong. Cloning and Functional Analysis of Gynostemma pentaphyllum GpMIR156a and GpMIR166b [J]. Biotechnology Bulletin, 2022, 38(7): 186-193. |
| [6] | CHEN Jia-min, LIU Yong-jie, MA Jin-xiu, LI Dan, GONG Jie, ZHAO Chang-ping, GENG Hong-wei, GAO Shi-qing. Expression Pattern Analysis of Histone Methyltransferase Under Drought Stress in Hybrid Wheat [J]. Biotechnology Bulletin, 2022, 38(7): 51-61. |
| [7] | LIU Jing-jing, LIU Xiao-rui, LI Lin, WANG Ying, YANG Hai-yuan, DAI Yi-fan. Establishment of Porcine Fetal Fibroblasts with OXTR-knockout Using CRISPR/Cas9 [J]. Biotechnology Bulletin, 2022, 38(6): 272-278. |
| [8] | WANG Nan, ZHANG Rui, PAN Yang-yang, HE Hong-hong, WANG Jing-lei, CUI Yan, YU Si-jiu. Cloning of Bos grunniens TGF-β1 Gene and Its Expression in Major Organs of Female Reproductive System [J]. Biotechnology Bulletin, 2022, 38(6): 279-290. |
| [9] | LI Yu-hang, WANG Xing-ping, YANG Jian, LUORENG Zhuo-ma, REN Qian-qian, WEI Da-wei, MA Yun. Expression and Functional Analysis of miR-665 in Bovine Mammary Epithelial Cell Inflammation [J]. Biotechnology Bulletin, 2022, 38(5): 159-168. |
| [10] | LI Yang, ZHANG Xiao-tian, PIAO Jing-zi, ZHOU Ru-jun, LI Zi-bo, GUAN Hai-wen. Cloning and Bioinformatics Analysis of Blue-light Receptor EaWC 1 Gene in Elsinoë arachidis [J]. Biotechnology Bulletin, 2022, 38(5): 93-99. |
| [11] | ZHANG Lin, WEI Zhen-zhen, SONG Cheng-wei, GUO Li-li, GUO Qi, HOU Xiao-gai, WANG Hua-fang. Cloning and Expression Analysis of PoFD Gene from Paeonia ostii ‘Fengdan’ [J]. Biotechnology Bulletin, 2022, 38(11): 104-111. |
| [12] | FAN Ya-peng, RUI Cun, ZHANG Yue-xin, CHEN Xiu-gui, LU Xu-ke, WANG Shuai, ZHANG Hong, XU Nan, WANG Jing, CHEN Chao, YE Wu-wei. Cloning,Expression and Preliminary Bioinformatics Analysis of the Alkaline Tolerant Gene GhZAT12 in Gossypium hirsutum [J]. Biotechnology Bulletin, 2021, 37(8): 121-130. |
| [13] | DU Zhen-wei, ZHU Shuai-peng, MA Xiang-fei, LI Dong-hua, SUN Gui-rong. Cloning,Expression and Bioinformatics Analysis of the CDS Region of Chicken CEBPA Gene [J]. Biotechnology Bulletin, 2021, 37(8): 203-212. |
| [14] | HAO Xiang-yang, LIU Fan, WU Huan, WANG Bin, SUN Xue-li, XIANG Lei-lei, WANG Tian-chi, LAI Zhong-xiong, CHENG Chun-zhen. Cloning and Expression Analysis of GjPAL Genes in Gerbera jamesonni [J]. Biotechnology Bulletin, 2021, 37(6): 13-23. |
| [15] | HAN Zhan-hong, ZONG Yuan-yuan, ZHANG Xue-mei, WANG Bin, PRUSKY Dov, BI Yang. Bioinformatics,Subcellular Localization and Expression Analysis of erg4 in Penicillium expansum [J]. Biotechnology Bulletin, 2021, 37(12): 60-70. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||