








Biotechnology Bulletin ›› 2024, Vol. 40 ›› Issue (4): 97-109.doi: 10.13560/j.cnki.biotech.bull.1985.2023-1025
Previous Articles Next Articles
LI Xing-rong1,2( ), TAN Zhi-bing3, ZHAO Yan3, LI Yao-kui2, ZHAO Bing-ran1,2(
), TAN Zhi-bing3, ZHAO Yan3, LI Yao-kui2, ZHAO Bing-ran1,2( ), TANG Li1,2(
), TANG Li1,2( )
)
Received:2023-11-02
Online:2024-04-26
Published:2024-04-30
Contact:
ZHAO Bing-ran, TANG Li
E-mail:L3141429315@163.com;brzhao652@hhrrc.ac.cn;tangli@hhrrc.ac.cn
LI Xing-rong, TAN Zhi-bing, ZHAO Yan, LI Yao-kui, ZHAO Bing-ran, TANG Li. Cloning and Functional Analysis of OsLCT3, a Low-affinity Cation Transporter Gene of Rice[J]. Biotechnology Bulletin, 2024, 40(4): 97-109.
| 引物名称 Primer name | 引物序列 Primer sequence(5'-3') | 引物用途 Primer usage |
|---|---|---|
| T3-CDS-F | TATCTTATGGACTGGTCTGTAGC | OsLCT3 CDS扩增、测序 |
| T3-CDS-R | TCTGTTGTCTGATGATGATTTTG | OsLCT3 CDS amplification and sequencing |
| T3-C-F | ATGGTGGCCCTTTCTATCCA | OsLCT3 RACE核心序列扩增 |
| T3-C-R | TCACTGTATCTCCAAAACCA | OsLCT3 RACE core sequence amplification |
| T3-3-F2 | TTGCCTCTGCAATAGTTGCC | OsLCT3 3' RACE 扩增 |
| T3-3-F3 | GTACTGTAATGGTTACACTTAG | OsLCT3 3' RACE amplification |
| T3-5-R2 | GAGGAAATACCAAGCCACCATC | OsLCT3 5' RACE 扩增 |
| T3-5-R3 | CGAAGTGATACATCCCTCAACCTG | OsLCT3 5' RACE amplification |
| OsLCT3-F2 | ATGGTGGCTTGGTATTTCCTC | OsLCT3 DNA片段扩增 |
| OsLCT3-R2 | CACCTCATTGGCATTACTGTCC | Amplification of OsLCT3 DNA fragments |
| OsLCT3-F3 | GAGATCCGTGCGAAAGTGCT | OsLCT3 DNA片段扩增 Amplification of OsLCT3 DNA fragments |
| OsLCT3-R3 | AGAGTGAGTGAAAAGCCCGA | |
| OsActin1-F | TGCTATGTACGTCGCCATCCAG | OsActin1 DNA片段扩增 |
| OsActin1-R | AATGAGTAACCACGCTCCGTCA | Amplification of OsActin1 DNA fragments |
| T3-qP-F | GGTGGTTCTGGGAAGACTAAA | OsLCT3 RT-qPCR分析 |
| T3-qP-R | ATGGAGAGCAGACCTGATATTG | OsLCT3 RT-qPCR analysis |
| OsActin1-qP-F | CAACACCCCTGCTATGTACG | OsActin1 RT-qPCR分析 |
| OsActin1-qP-R | CATCACCAGAGTCCAACACAA | OsActin1 RT-qPCR analysis |
| T3P1-F | GGCAGTACATTGGAAGCTTGTCTG | OsLCT3 敲除载体靶位点1 接头引物 |
| T3P1-R | AAACCAGACAAGCTTCCAATGTAC | OsLCT3 knockout vector target site 1 adaptor primer |
| T3P2-F | GCCGTCAATTACAACGCCACCGG | OsLCT3 敲除载体靶位点2 接头引物 |
| T3P2-R | AAACCCGGTGGCGTTGTAATTGA | OsLCT3 knockout vector target site 2 adapter primers |
| T3-p1132-F | cgcggtggcggccgctctagaATGGTGGCCCTTTCTATCCAT | OsLCT3亚细胞定位 |
| T3-p1132-R | gataagcttgatatcgaattcCTGTATCTCCAAAACCACCGCG | OsLCT3 subcellular localization |
| pYES2-T3-F | gggaatattaagcttggtaccATGGTGGCCCTTTCTATCCAT | OsLCT3酵母载体构建 |
| YES2-T3-R | gatggatatctgcagaattcTCACTGTATCTCCAAAACCACCG | OsLCT3 yeast vector construction |
| HPT-F | GCTCCATACAAGCCAACCACG | Hpt片段扩增 |
| HPT-R | CCTGCCTGAAACCGAACTGC | Hpt fragment amplification |
| CAS9-F | CGAGACGAACGGTGAGACTGGTG | Cas9片段扩增 |
| CAS9-R | GGTGCTTGTTGTAGGCGGAGAGG | Cas9 fragment amplification |
| OsLCT3-CAS-F | CTGCGGTATATCATCCCAATGCT | OsLCT3的靶点PCR扩增 |
| OsLCT3-CAS-R | CAACTATGGTGAAAACTTCGGCG | PCR amplification of OsLCT3 targets |
| pYES2-T3-eGFP-F | gggaatattaagcttggtaccATGGTGGCCCTTTCTATCCAT | OsLCT3-eGFP酵母载体构建 |
| pYES2-T3-eGFP-R | tgatggatatctgcagaattcTTACTTGTACAGCTCGTCCATGCC | OsLCT3-eGFP yeast vector construction |
Table 1 Primers and sequences used in this study
| 引物名称 Primer name | 引物序列 Primer sequence(5'-3') | 引物用途 Primer usage |
|---|---|---|
| T3-CDS-F | TATCTTATGGACTGGTCTGTAGC | OsLCT3 CDS扩增、测序 |
| T3-CDS-R | TCTGTTGTCTGATGATGATTTTG | OsLCT3 CDS amplification and sequencing |
| T3-C-F | ATGGTGGCCCTTTCTATCCA | OsLCT3 RACE核心序列扩增 |
| T3-C-R | TCACTGTATCTCCAAAACCA | OsLCT3 RACE core sequence amplification |
| T3-3-F2 | TTGCCTCTGCAATAGTTGCC | OsLCT3 3' RACE 扩增 |
| T3-3-F3 | GTACTGTAATGGTTACACTTAG | OsLCT3 3' RACE amplification |
| T3-5-R2 | GAGGAAATACCAAGCCACCATC | OsLCT3 5' RACE 扩增 |
| T3-5-R3 | CGAAGTGATACATCCCTCAACCTG | OsLCT3 5' RACE amplification |
| OsLCT3-F2 | ATGGTGGCTTGGTATTTCCTC | OsLCT3 DNA片段扩增 |
| OsLCT3-R2 | CACCTCATTGGCATTACTGTCC | Amplification of OsLCT3 DNA fragments |
| OsLCT3-F3 | GAGATCCGTGCGAAAGTGCT | OsLCT3 DNA片段扩增 Amplification of OsLCT3 DNA fragments |
| OsLCT3-R3 | AGAGTGAGTGAAAAGCCCGA | |
| OsActin1-F | TGCTATGTACGTCGCCATCCAG | OsActin1 DNA片段扩增 |
| OsActin1-R | AATGAGTAACCACGCTCCGTCA | Amplification of OsActin1 DNA fragments |
| T3-qP-F | GGTGGTTCTGGGAAGACTAAA | OsLCT3 RT-qPCR分析 |
| T3-qP-R | ATGGAGAGCAGACCTGATATTG | OsLCT3 RT-qPCR analysis |
| OsActin1-qP-F | CAACACCCCTGCTATGTACG | OsActin1 RT-qPCR分析 |
| OsActin1-qP-R | CATCACCAGAGTCCAACACAA | OsActin1 RT-qPCR analysis |
| T3P1-F | GGCAGTACATTGGAAGCTTGTCTG | OsLCT3 敲除载体靶位点1 接头引物 |
| T3P1-R | AAACCAGACAAGCTTCCAATGTAC | OsLCT3 knockout vector target site 1 adaptor primer |
| T3P2-F | GCCGTCAATTACAACGCCACCGG | OsLCT3 敲除载体靶位点2 接头引物 |
| T3P2-R | AAACCCGGTGGCGTTGTAATTGA | OsLCT3 knockout vector target site 2 adapter primers |
| T3-p1132-F | cgcggtggcggccgctctagaATGGTGGCCCTTTCTATCCAT | OsLCT3亚细胞定位 |
| T3-p1132-R | gataagcttgatatcgaattcCTGTATCTCCAAAACCACCGCG | OsLCT3 subcellular localization |
| pYES2-T3-F | gggaatattaagcttggtaccATGGTGGCCCTTTCTATCCAT | OsLCT3酵母载体构建 |
| YES2-T3-R | gatggatatctgcagaattcTCACTGTATCTCCAAAACCACCG | OsLCT3 yeast vector construction |
| HPT-F | GCTCCATACAAGCCAACCACG | Hpt片段扩增 |
| HPT-R | CCTGCCTGAAACCGAACTGC | Hpt fragment amplification |
| CAS9-F | CGAGACGAACGGTGAGACTGGTG | Cas9片段扩增 |
| CAS9-R | GGTGCTTGTTGTAGGCGGAGAGG | Cas9 fragment amplification |
| OsLCT3-CAS-F | CTGCGGTATATCATCCCAATGCT | OsLCT3的靶点PCR扩增 |
| OsLCT3-CAS-R | CAACTATGGTGAAAACTTCGGCG | PCR amplification of OsLCT3 targets |
| pYES2-T3-eGFP-F | gggaatattaagcttggtaccATGGTGGCCCTTTCTATCCAT | OsLCT3-eGFP酵母载体构建 |
| pYES2-T3-eGFP-R | tgatggatatctgcagaattcTTACTTGTACAGCTCGTCCATGCC | OsLCT3-eGFP yeast vector construction |
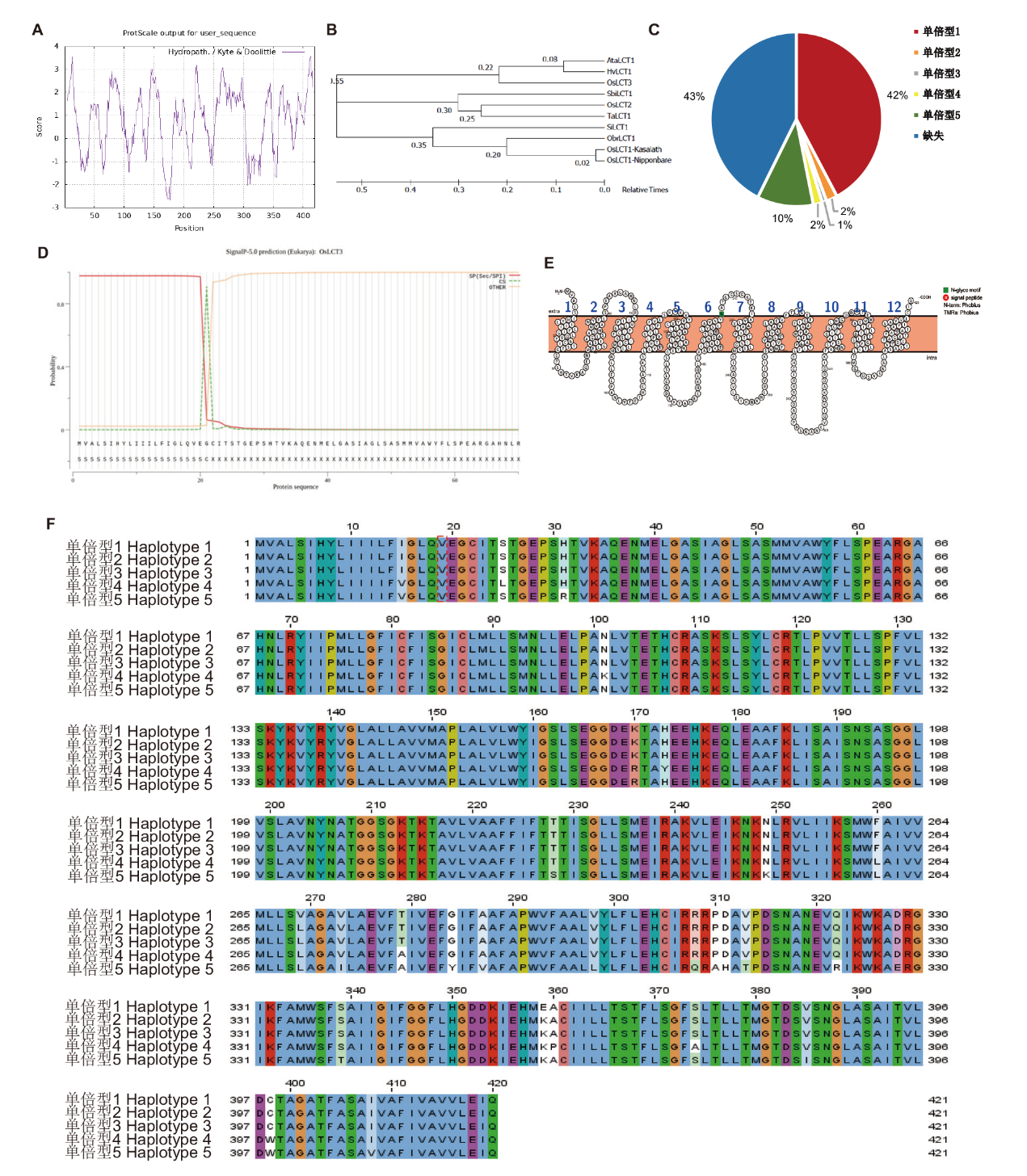
Fig. 2 Bioinformatics analysis for OsLCT3 A: Hydrophobicity of OsLCT3 protein. B: Phylogenetic analysis of LCT family members. C: Distribution frequency of the OsLCT3 haplotype. D: Signaling peptide prediction of OsLCT3 protein. E: OsLCT3 transmembrane domain analysis. F: OsLCT3 haplotype analysis. Ta: Triticum aestivum; Sbi: Sorghum bicolor; Os: Oryza sativa; Si: Setaria italica; Ata: Aegilops tauschii; Hv: Hordeum vulgare; Obr: Oryza brachyantha
| 蛋白质 Protein | 氨基酸数 Amino acid No./aa | 分子式 Formula | 分子量 Molecular weight/kD | 等电点 pI | 不稳定指数 Instability index | 平均疏水性 Hydropathy index | 正 / 负电残基数 Acid-base amino acid | 脂溶指数 Aliphatic index |
|---|---|---|---|---|---|---|---|---|
| OsLCT3 | 420 | C2088H3315N511O567S22 | 45.355 | 6.51 | 25.72 | 0.777 | 28/30 | 121.95 |
Table 2 Physicochemical properties of OsLCT3 protein
| 蛋白质 Protein | 氨基酸数 Amino acid No./aa | 分子式 Formula | 分子量 Molecular weight/kD | 等电点 pI | 不稳定指数 Instability index | 平均疏水性 Hydropathy index | 正 / 负电残基数 Acid-base amino acid | 脂溶指数 Aliphatic index |
|---|---|---|---|---|---|---|---|---|
| OsLCT3 | 420 | C2088H3315N511O567S22 | 45.355 | 6.51 | 25.72 | 0.777 | 28/30 | 121.95 |
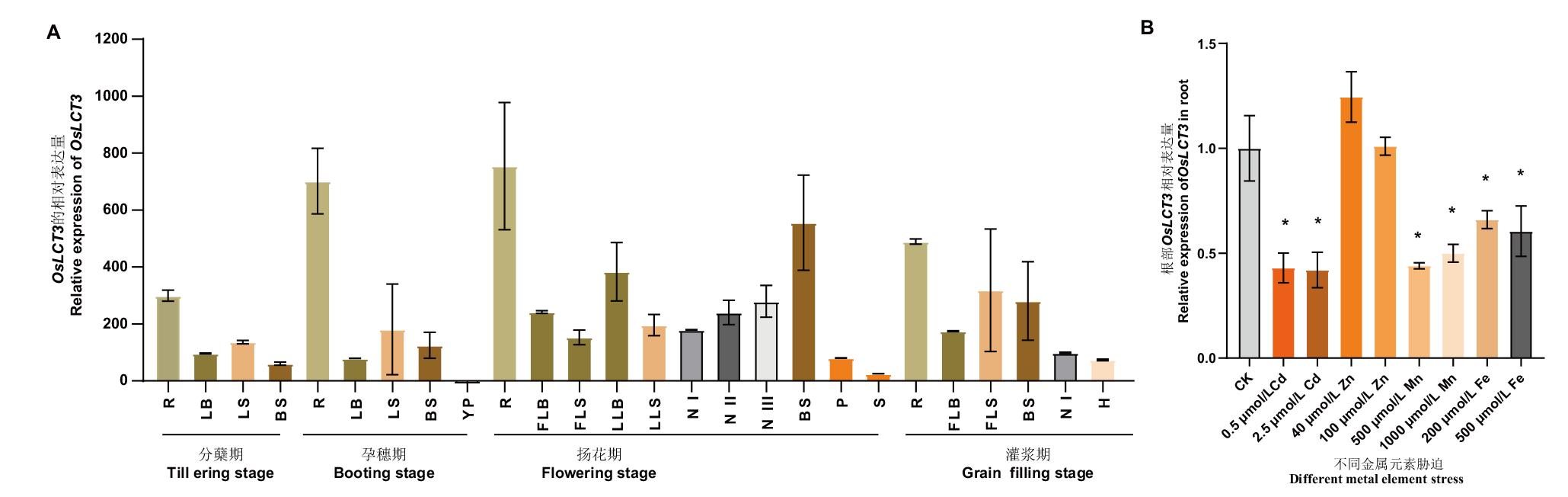
Fig. 3 Expression patterns of OsLCT3 A: Expression analysis of OsLCT3 at different growth stages. The expression level of OsLCT3 in young panicle of booting stage was set to 1, and the 2-△△ Ct method was used to estimate the relative expression of OsLCT3 in each tissue at each stage(R: Root. LB: Leaf blade. LS: Leaf sheath. BS: Basal stem. YP: Young panicle. FLB: Flag leaf blade. FLS: Flag leaf sheath. LLB: Lower leaf blade. LLS: Lower leaf sheath. N I: Node I. N II: Node II. N III: Node III. P: Peduncle. S: Spikelet. H: Husk). B: Response of OsLCT3 to stress of different metal elements for 7 d. Standard deviation is based on three biological replicates. One-way ANOVA was used for data analysis,* indicating a significant difference at the level of 0.05 compared with the control by student’s t- test

Fig. 4 Subcellular localization of OsLCT3 eGFP: Enhanced green fluorescence. mCherry: Red fluorescent protein. Bright field: Bright field. Merge: Fluorescence integration. Scale bar=10 μm
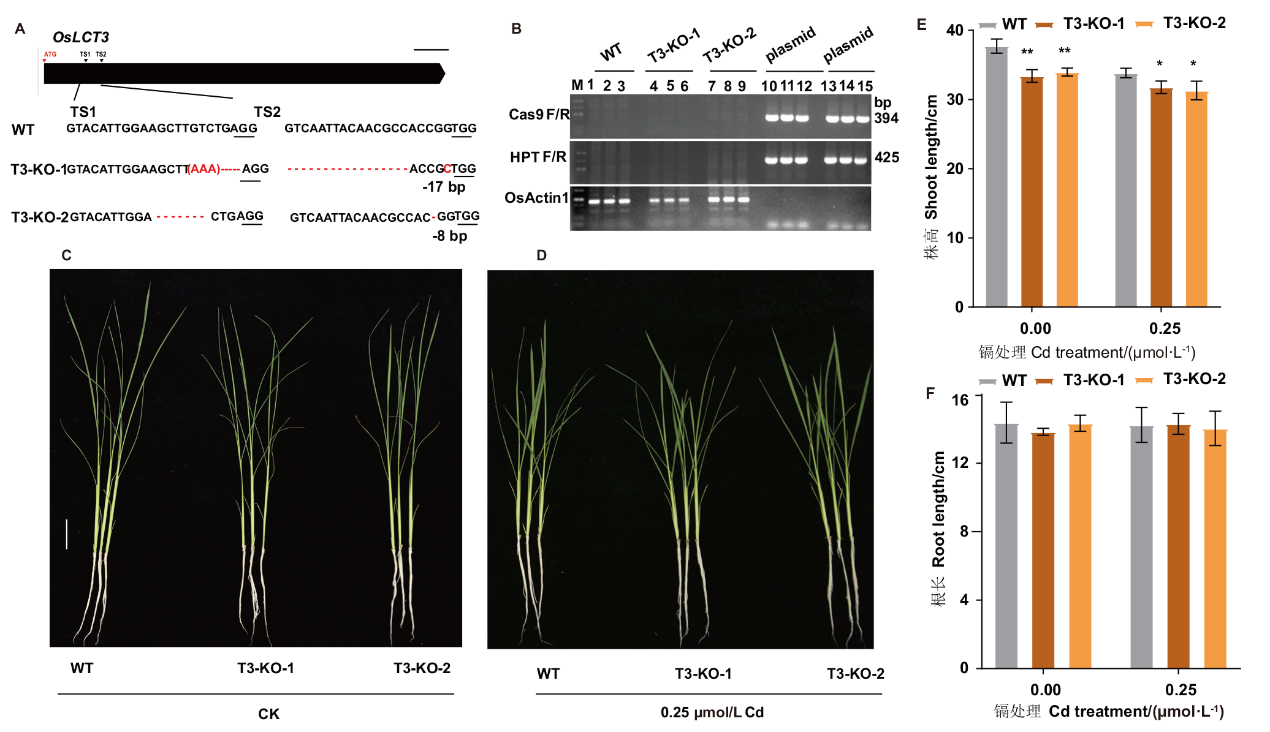
Fig. 5 Growth of oslct3 and WT in hydroponic growth A-B: Identification of oslct3 homozygous mutants. C-D: Phenotypes of WT and knockout lines. E: Plant height. F: Root length. * or ** indicate statistically significant difference in comparison to WT at P< 0.05 or P< 0.01 by student’s t test, the same applies hereinafter. Scale bar=5 cm
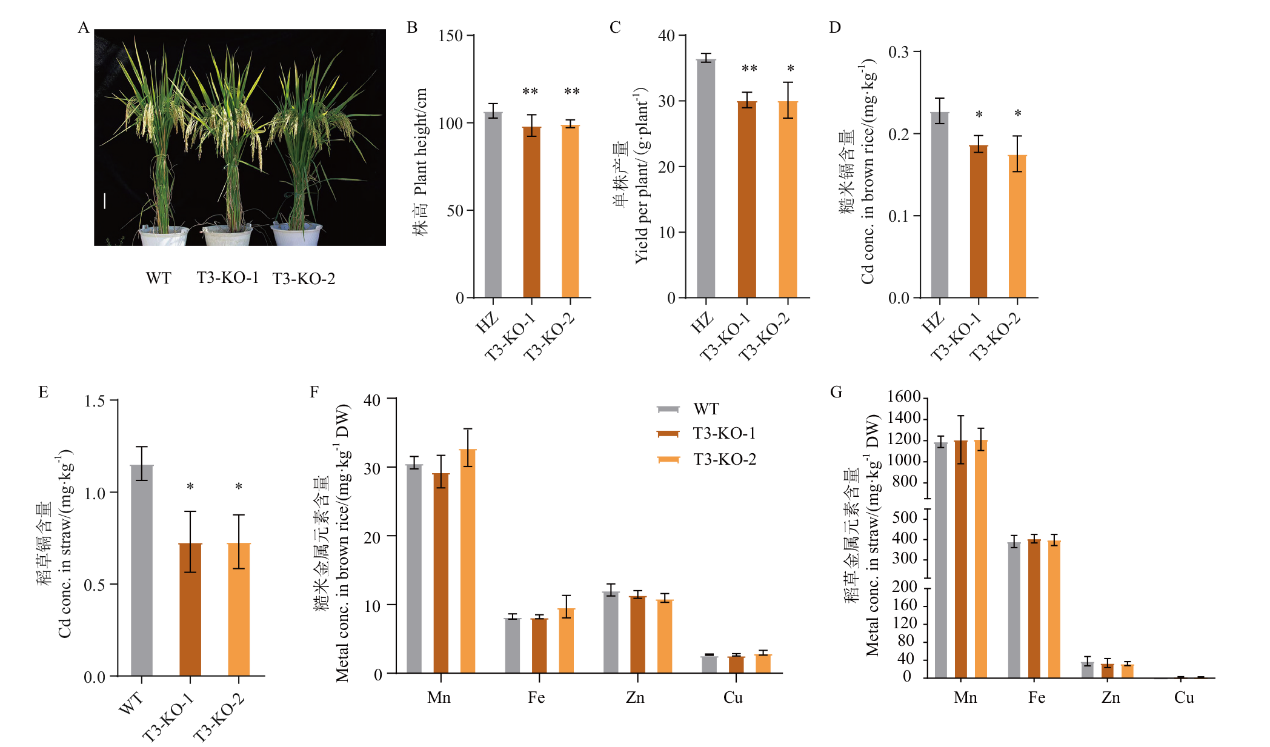
Fig. 7 Grain yield of oslct3 and metal contents in straw and brown rice under Cd-contaminated fields A: Plant phenotype at maturity. B: Plant height. C: Yield per plant. D-E: Cd content in brown rice and straw. F-G: Brown rice and straw metal element content. Scale bar=10 cm

Fig. 8 Identification of effects of OsLCT3 on the tolerance of yeast to Cd stress A: OsLCT3 Cd tolerance phenotypic identification in yeast. B: Cd tolerance growth curve analysis of OsLCT3 in yeast
| [1] |
Jamers A, Blust R, De Coen W, et al. An omics based assessment of cadmium toxicity in the green alga Chlamydomonas reinhardtii[J]. Aquat Toxicol, 2013, 126: 355-364.
doi: 10.1016/j.aquatox.2012.09.007 pmid: 23063003 |
| [2] | 汪鹏, 王静, 陈宏坪, 等. 我国稻田系统镉污染风险与阻控[J]. 农业环境科学学报, 2018, 37(7): 1409-1417. |
| Wang P, Wang J, Chen HP, et al. Cadmium risk and mitigation in paddy systems in China[J]. J Agro Environ Sci, 2018, 37(7): 1409-1417. | |
| [3] |
Clemens S, Ma JF. Toxic heavy metal and metalloid accumulation in crop plants and foods[J]. Annu Rev Plant Biol, 2016, 67: 489-512.
doi: 10.1146/annurev-arplant-043015-112301 pmid: 27128467 |
| [4] | Tang L, Li Y, Peng Y, et al. Breeding for rice cultivars with low cadmium accumulation[M]//Heavy Metal Toxicity and Tolerance in Plants: A Biological, Omics, and Genetic Engineering Approach. Chichester: John Wiley & Sons Ltd, 2023: 335-347. |
| [5] |
冀中英, 李曜魁, 孟前程, 等. 水稻积累及耐受镉和砷的分子机制与育种实践[J]. 植物遗传资源学报, 2023, 24(1): 75-85.
doi: 10.13430/j.cnki.jpgr.20220504002 |
| Ji ZY, Li YK, Meng QC, et al. Molecular mechanisms and breeding practices of accumulation and tolerance to cadmium and arsenic in rice[J]. J Plant Genet Resour, 2023, 24(1): 75-85. | |
| [6] |
Uraguchi S, Kamiya T, Clemens S, et al. Characterization of OsLCT1, a cadmium transporter from indica rice(Oryza sativa)[J]. Physiol Plant, 2014, 151(3): 339-347.
doi: 10.1111/ppl.12189 pmid: 24627964 |
| [7] |
Uraguchi S, Kamiya T, Sakamoto T, et al. Low-affinity cation transporter(OsLCT1)regulates cadmium transport into rice grains[J]. Proc Natl Acad Sci USA, 2011, 108(52): 20959-20964.
doi: 10.1073/pnas.1116531109 pmid: 22160725 |
| [8] | Kumar A. 水稻籽粒锌含量的全基因组关联分析及候选基因验证[D]. 北京: 中国农业科学院, 2021. |
| Kumar A. Genome-wide association analysis and validation of candidate genes[D]. Beijing: Chinese Academy of Agricultural Sciences, 2021. | |
| [9] | 李曜魁, 唐丽, 毛毕刚, 等. 籼稻低亲和阳离子转运蛋白基因OsLCT2的克隆与生物信息学分析[J]. 分子植物育种, 2016, 14(5): 1067-1074. |
| Li YK, Tang L, Mao BG, et al. Cloning and bioinformatics analysis of low-affinity cation transporter gene OsLCT2 in indica rice(Oryza sativa)[J]. Mol Plant Breed, 2016, 14(5): 1067-1074. | |
| [10] |
Tang L, Dong JY, Tan LT, et al. Overexpression of OsLCT2, a low-affinity cation transporter gene, reduces cadmium accumulation in shoots and grains of rice[J]. Rice, 2021, 14: 89.
doi: 10.1186/s12284-021-00530-8 pmid: 34693475 |
| [11] |
Schachtman DP, Kumar R, Schroeder JI, et al. Molecular and functional characterization of a novel low-affinity cation transporter(LCT1)in higher plants[J]. Proc Natl Acad Sci USA, 1997, 94(20): 11079-11084.
pmid: 9380762 |
| [12] |
Clemens S, Antosiewicz DM, Ward JM, et al. The plant cDNA LCT1 mediates the uptake of calcium and cadmium in yeast[J]. Proc Natl Acad Sci USA, 1998, 95(20): 12043-12048.
doi: 10.1073/pnas.95.20.12043 pmid: 9751787 |
| [13] |
Amtmann A, Fischer M, Marsh EL, et al. The wheat cDNA LCT1 generates hypersensitivity to sodium in a salt-sensitive yeast strain[J]. Plant Physiol, 2001, 126(3): 1061-1071.
pmid: 11457957 |
| [14] |
Antosiewicz DM, Hennig J. Overexpression of LCT1 in tobacco enhances the protective action of calcium against cadmium toxicity[J]. Environ Pollut, 2004, 129(2): 237-245.
doi: 10.1016/j.envpol.2003.10.025 pmid: 14987809 |
| [15] |
Wojas S, Ruszczyńska A, Bulska E, et al. Ca2+-dependent plant response to Pb2+ is regulated by LCT1[J]. Environ Pollut, 2007, 147(3): 584-592.
doi: 10.1016/j.envpol.2006.10.012 pmid: 17140712 |
| [16] |
Xia RZ, Zhou J, Cui HB, et al. Nodes play a major role in cadmium(Cd)storage and redistribution in low-Cd-accumulating rice(Oryza sativa L.) cultivars[J]. Sci Total Environ, 2023, 859: 160436.
doi: 10.1016/j.scitotenv.2022.160436 URL |
| [17] | 邱冠凯, 倪大虎, 韩忠民, 等. 水稻铁吸收与转运机理[J]. 土壤与作物, 2022, 11(2): 170-178. |
| Qiu GK, Ni DH, Han ZM, et al. Mechanisms of iron absorption and transport in rice: a review[J]. Soils Crops, 2022, 11(2): 170-178. | |
| [18] |
Tan LT, Qu MM, Zhu YX, et al. ZINC TRANSPORTER5 and ZINC TRANSPORTER9 function synergistically in zinc/cadmium uptake[J]. Plant Physiol, 2020, 183(3): 1235-1249.
doi: 10.1104/pp.19.01569 pmid: 32341004 |
| [19] |
薛欣月, 于雪然, 刘晓刚, 等. 水稻锌吸收、转运、累积机理研究进展[J]. 生物技术通报, 2022, 38(4): 29-43.
doi: 10.13560/j.cnki.biotech.bull.1985.2020-1484 |
| Xue XY, Yu XR, Liu XG, et al. Research progress in absorption, transportation and accumulation mechanism of zinc in rice[J]. Biotechnol Bull, 2022, 38(4): 29-43. | |
| [20] |
王璐瑶, 陈謇, 赵守清, 等. 水稻镉积累特性的生理和分子机制研究概述[J]. 植物学报, 2022, 57(2): 236-249.
doi: 10.11983/CBB21222 |
| Wang LY, Chen J, Zhao SQ, et al. Research progress of the physiological and molecular mechanisms of cadmium accumulation in rice[J]. Chin Bull Bot, 2022, 57(2): 236-249. | |
| [21] |
Tan LT, Zhu YX, Fan T, et al. OsZIP7 functions in xylem loading in roots and inter-vascular transfer in nodes to deliver Zn/Cd to grain in rice[J]. Biochem Biophys Res Commun, 2019, 512(1): 112-118.
doi: 10.1016/j.bbrc.2019.03.024 URL |
| [22] |
Takahashi R, Ishimaru Y, Shimo H, et al. The OsHMA2 transporter is involved in root-to-shoot translocation of Zn and Cd in rice[J]. Plant Cell Environ, 2012, 35(11): 1948-1957.
doi: 10.1111/pce.2012.35.issue-11 URL |
| [23] |
Hao XH, Zeng M, Wang J, et al. A node-expressed transporter OsCCX2 is involved in grain cadmium accumulation of rice[J]. Front Plant Sci, 2018, 9: 476.
doi: 10.3389/fpls.2018.00476 pmid: 29696032 |
| [24] | Luo JS, Huang J, Zeng DL, et al. A defensin-like protein drives cadmium efflux and allocation in rice[J]. Nat Commun, 2018, 9(1): 645. |
| [1] | YANG Qi, WEI Zi-di, SONG Juan, TONG Kun, YANG Liu, WANG Jia-han, LIU Hai-yan, LUAN Wei-jiang, MA Xuan. Construction and Transcriptomic Analysis of Rice Histone H1 Triple Mutant [J]. Biotechnology Bulletin, 2024, 40(4): 85-96. |
| [2] | LIU Jia-ning, LI Meng, YANG Xin-sen, WU Wei, PEI Xin-wu, YUAN Qian-hua. Impact of Different Water Management Cultivation Methods on the Rhizosphere Bacteria Community of Shanlan Upland Rice [J]. Biotechnology Bulletin, 2024, 40(3): 242-250. |
| [3] | LI Xue, LI Rong-ou, KONG Mei-yi, HUANG Lei. The Growth Promoting Effect of Bacillus amyloliquefaciens SQ-2 on Rice [J]. Biotechnology Bulletin, 2024, 40(2): 109-119. |
| [4] | ZHAO Yao, WEN Lang, LUO Shao-dan, LI Zi-xing, LIU Chao-chao. Identification of HMA Gene Family and Cadmium Transport Function of SlHMA1 in Tomato [J]. Biotechnology Bulletin, 2024, 40(2): 212-222. |
| [5] | ZHANG Chao, WANG Zi-rui, SUN Ya-li, MAO Xin-chen, TANG Jia-qi, YU Heng-xiu. Functional Study of Vitamin B1 Synthesis-related Gene OsTHIC in Rice [J]. Biotechnology Bulletin, 2024, 40(2): 99-108. |
| [6] | LIN Xin-yan, ZHANG Chuan-zhong, DAI Bing, WANG Xin-heng, LIU Jian-feng, WEN Li, XU Xing-jian, FANG Jun. Advances in Genetic and Molecular Mechanisms of Pre-harvest Sprouting in Rice [J]. Biotechnology Bulletin, 2024, 40(1): 24-31. |
| [7] | WANG Zi-ying, LONG Chen-jie, FAN Zhao-yu, ZHANG Lei. Screening of OsCRK5-interacted Proteins in Rice Using Yeast Two-hybrid System [J]. Biotechnology Bulletin, 2023, 39(9): 117-125. |
| [8] | WU Yuan-ming, LIN Jia-yi, LIU Yu-xi, LI Dan-ting, ZHANG Zong-qiong, ZHENG Xiao-ming, PANG Hong-bo. Identification of Rice Plant Height-associated QTL Using BSA-seq and RNA-seq [J]. Biotechnology Bulletin, 2023, 39(8): 173-184. |
| [9] | YAO Sha-sha, WANG Jing-jing, WANG Jun-jie, LIANG Wei-hong. Molecular Mechanisms of Rice Grain Size Regulation Related to Plant Hormone Signaling Pathways [J]. Biotechnology Bulletin, 2023, 39(8): 80-90. |
| [10] | LI Yu, LI Su-zhen, CHEN Ru-mei, LU Hai-qiang. Advances in the Regulation of Iron Homeostasis by bHLH Transcription Factors in Plant [J]. Biotechnology Bulletin, 2023, 39(7): 26-36. |
| [11] | LIANG Cheng-gang, WANG Yan, LI Tian, OHSUGI Ryu, AOKI Naohiro. Effect of SP1 on Panicle Architecture by Regulating Carbohydrate Remobilization [J]. Biotechnology Bulletin, 2023, 39(5): 152-159. |
| [12] | ZHOU Ding-ding, LI Hui-hu, TANG Xing-yong, YU Fa-xin, KONG Dan-yu, LIU Yi. Research Progress in the Biosynthesis and Regulation of Glycyrrhizic Acid and Liquiritin [J]. Biotechnology Bulletin, 2023, 39(5): 44-53. |
| [13] | YANG Mao, LIN Yu-feng, DAI Yang-shuo, PAN Su-jun, PENG Wei-ye, YAN Ming-xiong, LI Wei, WANG Bing, DAI Liang-ying. OsDIS1 Negatively Regulates Rice Drought Tolerance Through Antioxidant Pathways [J]. Biotechnology Bulletin, 2023, 39(2): 88-95. |
| [14] | JIANG Min-xuan, LI Kang, LUO Liang, LIU Jian-xiang, LU Hai-ping. Advances on the Expressions of Foreign Proteins in Plants [J]. Biotechnology Bulletin, 2023, 39(11): 110-122. |
| [15] | JIANG Nan, SHI Yang, ZHAO Zhi-hui, LI Bin, ZHAO Yi-hui, YANG Jun-biao, YAN Jia-ming, JIN Yu-fan, CHEN Ji, HUANG Jin. Expression and Functional Analysis of OsPT1 Gene in Rice Under Cadmium Stress [J]. Biotechnology Bulletin, 2023, 39(1): 166-174. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||