








Biotechnology Bulletin ›› 2024, Vol. 40 ›› Issue (5): 191-202.doi: 10.13560/j.cnki.biotech.bull.1985.2023-1161
Previous Articles Next Articles
LI Jia-xin( ), LI Hong-yan, LIU Li-e, ZHANG Tian, ZHOU Wu(
), LI Hong-yan, LIU Li-e, ZHANG Tian, ZHOU Wu( )
)
Received:2023-12-10
Online:2024-05-26
Published:2024-06-13
Contact:
ZHOU Wu
E-mail:2544313591@qq.com;zhouwu870624@163.com
LI Jia-xin, LI Hong-yan, LIU Li-e, ZHANG Tian, ZHOU Wu. Identification and Expression Analysis of the NRAMP Family in Seabuckthorn Under Lead Stress[J]. Biotechnology Bulletin, 2024, 40(5): 191-202.
| 基因名称 Gene name | 基因ID Gene ID | 蛋白序列长度 Protein sequence length/aa | 分子量 Molecular weight/kD | 理论等电点 Isoelectric point | 亚细胞定位 Subcellular localization |
|---|---|---|---|---|---|
| HrLNRAMP1 | Hiprha1gene15634 | 512 | 56.21 | 4.89 | 质膜Plas |
| HrLNRAMP2 | Hiprha1gene30069 | 513 | 55.52 | 9.53 | 质膜Plas |
| HrLNRAMP3 | Hiprha1gene21177 | 626 | 69.13 | 8.50 | 质膜Plas |
| HrLNRAMP4 | Hiprha1gene09270 | 1 291 | 141.46 | 5.79 | 质膜Plas |
| HrLNRAMP5 | Hiprha1gene12503 | 530 | 58.39 | 4.93 | 质膜Plas |
| HrLNRAMP6 | Hiprha1gene10061 | 532 | 57.72 | 9.33 | 质膜Plas |
| HrLNRAMP7 | Hiprha1gene12853 | 538 | 58.68 | 5.94 | 质膜Plas |
| HrLNRAMP8 | Hiprha1gene11195 | 1 295 | 141.63 | 6.08 | 质膜Plas |
| HrLNRAMP9 | Hiprha1gene27351 | 513 | 56.33 | 4.73 | 质膜Plas |
| HrLNRAMP10 | Hiprha1gene30205 | 502 | 54.94 | 8.50 | 质膜Plas |
| HrLNRAMP11 | Hiprha1gene16210 | 327 | 36.24 | 6.40 | 液泡膜Vacu |
Table 1 Physicochemical properties and subcellular localization of seabuckthorn NRAMP protein
| 基因名称 Gene name | 基因ID Gene ID | 蛋白序列长度 Protein sequence length/aa | 分子量 Molecular weight/kD | 理论等电点 Isoelectric point | 亚细胞定位 Subcellular localization |
|---|---|---|---|---|---|
| HrLNRAMP1 | Hiprha1gene15634 | 512 | 56.21 | 4.89 | 质膜Plas |
| HrLNRAMP2 | Hiprha1gene30069 | 513 | 55.52 | 9.53 | 质膜Plas |
| HrLNRAMP3 | Hiprha1gene21177 | 626 | 69.13 | 8.50 | 质膜Plas |
| HrLNRAMP4 | Hiprha1gene09270 | 1 291 | 141.46 | 5.79 | 质膜Plas |
| HrLNRAMP5 | Hiprha1gene12503 | 530 | 58.39 | 4.93 | 质膜Plas |
| HrLNRAMP6 | Hiprha1gene10061 | 532 | 57.72 | 9.33 | 质膜Plas |
| HrLNRAMP7 | Hiprha1gene12853 | 538 | 58.68 | 5.94 | 质膜Plas |
| HrLNRAMP8 | Hiprha1gene11195 | 1 295 | 141.63 | 6.08 | 质膜Plas |
| HrLNRAMP9 | Hiprha1gene27351 | 513 | 56.33 | 4.73 | 质膜Plas |
| HrLNRAMP10 | Hiprha1gene30205 | 502 | 54.94 | 8.50 | 质膜Plas |
| HrLNRAMP11 | Hiprha1gene16210 | 327 | 36.24 | 6.40 | 液泡膜Vacu |

Fig. 2 Phylogenetic analysis of NRAMP proteins in H. rhamnoides, A. thaliana and O. sativa Different branch colors indicate different subfamilies. Red triangles are labeled with H. rhamnoides NRAMP family members, blue pentagrams are labeled with Arabidopsis thaliana NRAMP family members, and yellow circles are labeled with Oryza sativa NRAMP family members
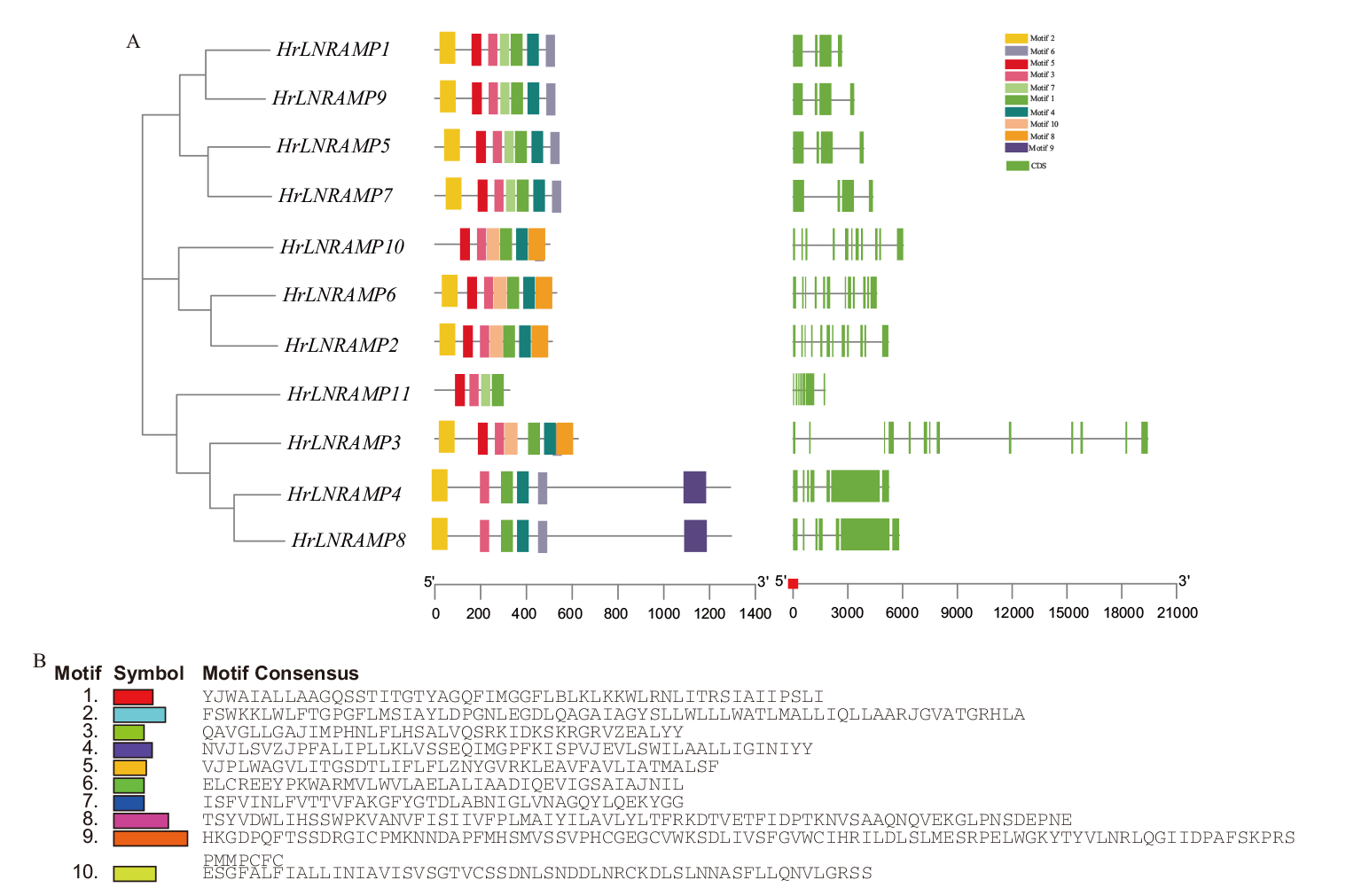
Fig. 4 NRAMP family exon-intron and motif analysis A: Structure and conserved motifs of the NRAMP family in seabuckthorn. B: Conserved motif composition of the NRAMP family in seabuckthorn
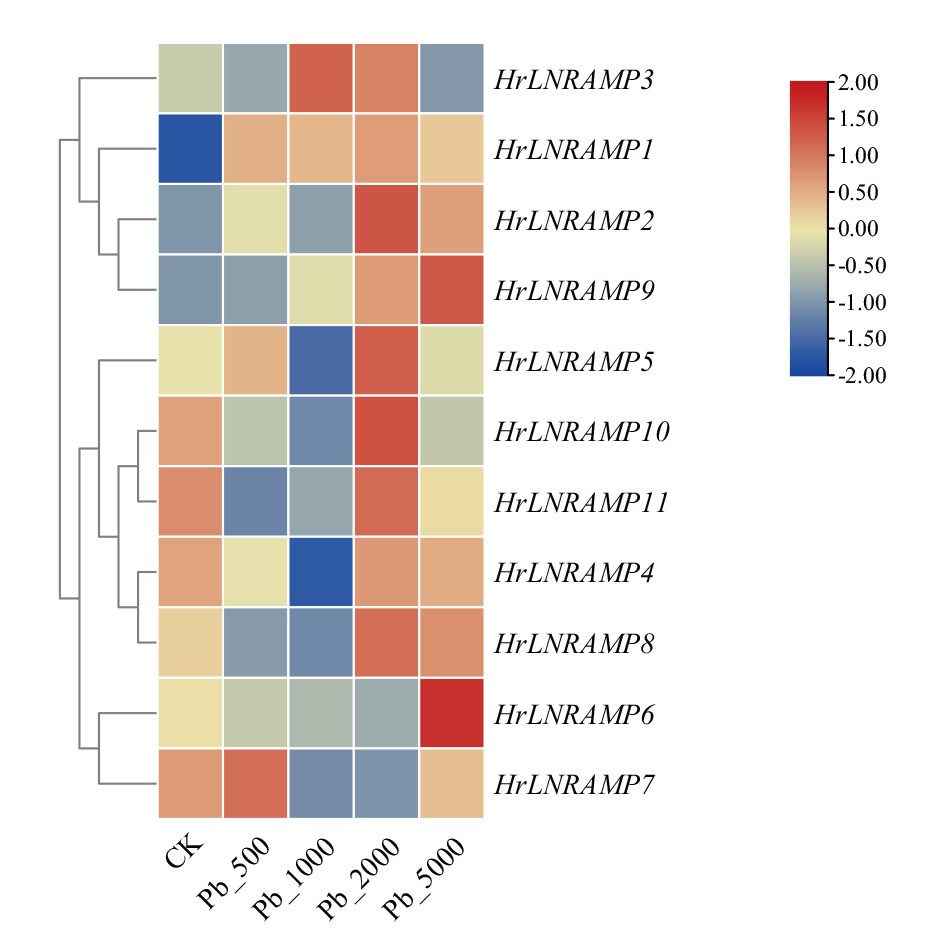
Fig. 6 Expression analysis of NRAMP gene in H. rhamnoi-des under different concentrations of lead stress CK: Control group, normal-growing seabuckthorn seedlings. Pb_500, Pb_1000, Pb_2000, Pb_5000: Experimental groups, with Pb stress concentrations of 500, 1 000, 2 000, and 5 000 mg/kg. The same below
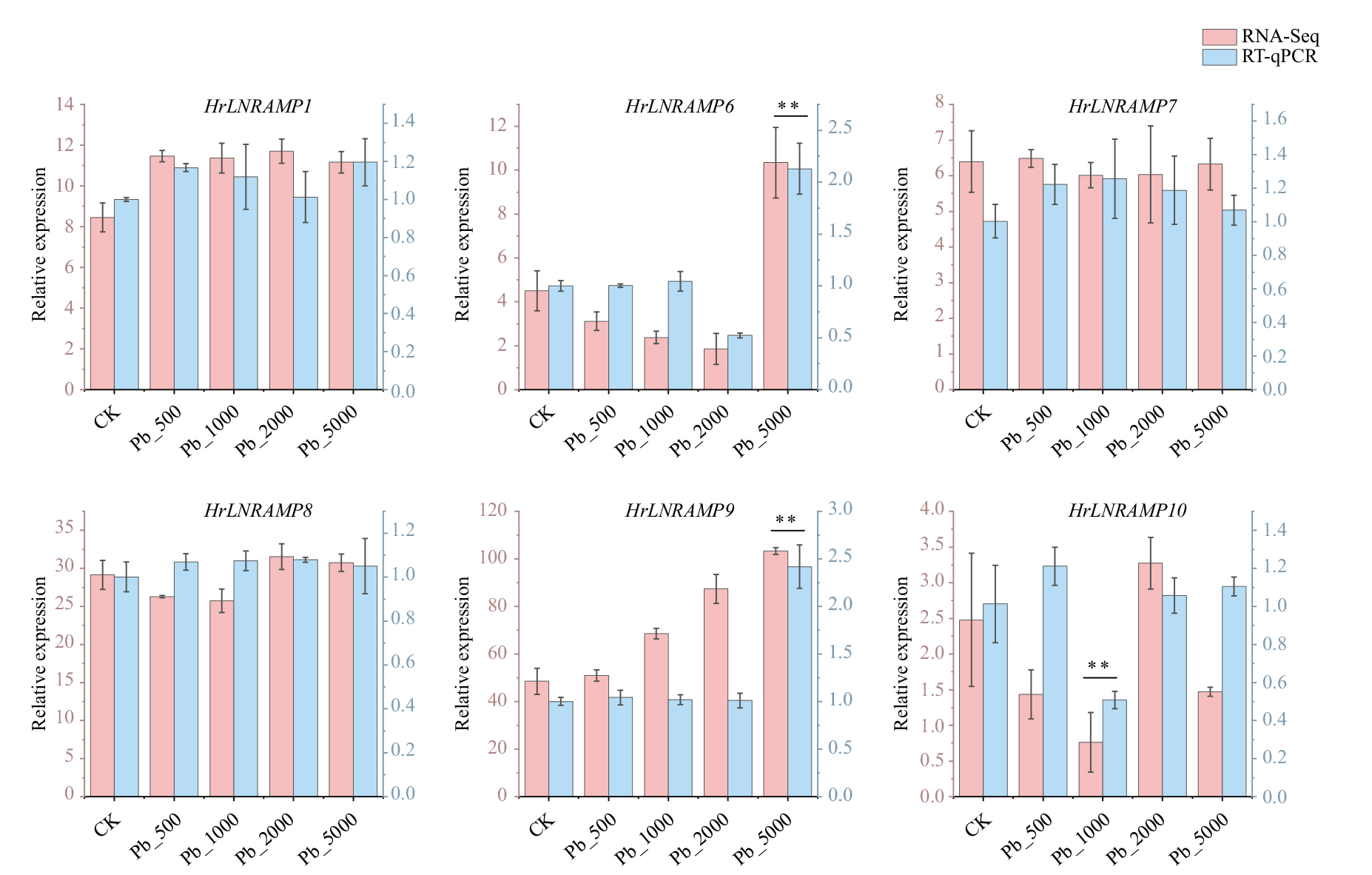
Fig. 7 Relative expressions of partial genes of HrLNRAMP gene family under lead stress conditions Data are means(±SD)of three biological replicates. Vertical bars indicate standard deviations. Asterisks indicate corresponding genes significantly upregulated or downregulated between the treatment and control(** P < 0.01)
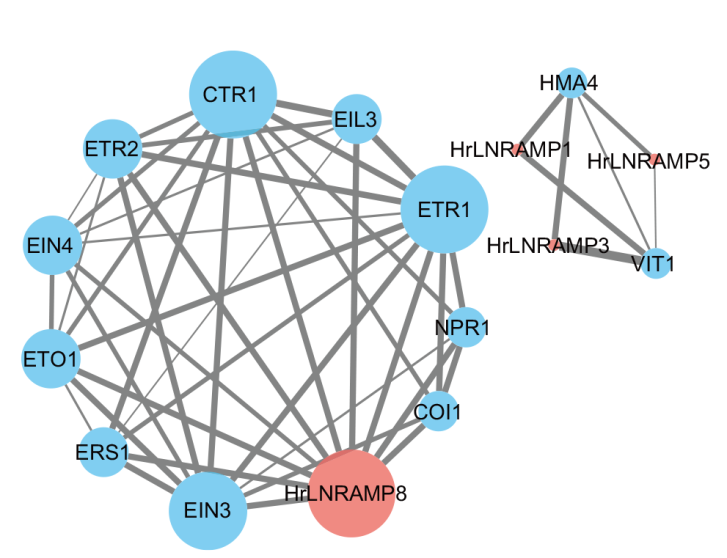
Fig. 10 Interaction protein network of HrLNRAMP geness in seabuckthorn Line thickness indicates the interaction strength; thicker lines denote stronger interactions, while thinner lines signify weaker ones. Blue denotes Arabidopsis proteins interacting with seabuckthorn, while red is specific to NRAMP proteins engaging in such interactions
| [1] | Nevo Y, Nelson N. The NRAMP family of metal-ion transporters[J]. Biochim Biophys Acta Mol Cell Res, 2006, 1763(7): 609-620. |
| [2] | Bozzi AT, Gaudet R. Molecular mechanism of NRAMP-family transition metal transport[J]. J Mol Biol, 2021, 433(16): 166991. |
| [3] |
Thomine S, Wang R, Ward JM, et al. Cadmium and iron transport by members of a plant metal transporter family in Arabidopsis with homology to NRAMP genes[J]. Proc Natl Acad Sci USA, 2000, 97(9): 4991-4996.
doi: 10.1073/pnas.97.9.4991 pmid: 10781110 |
| [4] | Gong ZH, Duan YQ, Liu DM, et al. Physiological and transcriptome analysis of response of soybean(Glycine max)to cadmium stress under elevated CO2 concentration[J]. J Hazard Mater, 2023, 448: 130950. |
| [5] | Tian WJ, He GD, Qin LJ, et al. Genome-wide analysis of the NRAMP gene family in potato(Solanum tuberosum): identification, expression analysis and response to five heavy metals stress[J]. Ecotoxicol Environ Saf, 2021, 208: 111661. |
| [6] | Fareed MM, Qasmi M, Shahid Z. The the structural, functional identification of natural resistance-associated macrophage protein as transporter elements in Oryza sativa[J]. European Journal of Volunteering and Community-based Projects, 2021, 1(2): 17-27. |
| [7] |
李一涵, 于浪柳, 李春燕, 等. 大麦NRAMP全基因组鉴定及重金属胁迫下基因表达分析[J]. 生物技术通报, 2022, 38(6): 103-111.
doi: 10.13560/j.cnki.biotech.bull.1985.2021-1006 |
| Li YH, Yu LL, Li CY, et al. Whole genome identification of barley NRAMP and gene expression analysis under heavy metal stress[J]. Biotechnol Bull, 2022, 38(6): 103-111. | |
| [8] | Hussain Q, Ye T, Shang CJ, et al. NRAMP gene family in Kandelia obovata: genome-wide identification, expression analysis, and response to five different copper stress conditions[J]. Front Plant Sci, 2024, 14: 1318383. |
| [9] | 陈可欣, 蒋贤达, 朱祝军, 等. 植物NRAMP家族参与金属离子吸收和分配的研究进展[J]. 植物生理学报, 2020, 56(3): 345-355. |
| Chen KX, Jiang XD, Zhu ZJ, et al. Advances in the study of plant NRAMP family involved in metal ion absorption and distribution[J]. Plant Physiol J, 2020, 56(3): 345-355. | |
| [10] | Kumar A, Singh G, Prasad B, et al. Genome wide analysis and identification of NRAMP gene family in wheat(Triticum aestivum L.)[J]. J Pharm Innov, 2022, 11(1): 499-504. |
| [11] |
Ihnatowicz A, Siwinska J, Meharg AA, et al. Conserved histidine of metal transporter AtNRAMP1 is crucial for optimal plant growth under manganese deficiency at chilling temperatures[J]. New Phytol, 2014, 202(4): 1173-1183.
doi: 10.1111/nph.12737 pmid: 24571269 |
| [12] | Żuchowski J. Phytochemistry and pharmacology of sea buckthorn(Elaeagnus rhamnoides; syn. Hippophae rhamnoides): progress from 2010 to 2021[J]. Phytochem Rev, 2023, 22(1): 3-33. |
| [13] | 徐智玮, 贾守宁, 赵国福, 等. 青海沙棘资源调查与产业发展建议[J]. 中国现代中药, 2020, 22(9): 1453-1457. |
| Xu ZW, Jia SN, Zhao GF, et al. Resources investigation and industrial development of Hippophae rhamnoides in Qinghai Province[J]. Mod Chin Med, 2020, 22(9): 1453-1457. | |
| [14] | Yu LY, Diao SF, Zhang GY, et al. Genome sequence and population genomics provide insights into chromosomal evolution and phytochemical innovation of Hippophae rhamnoides[J]. Plant Biotechnol J, 2022, 20(7): 1257-1273. |
| [15] |
Mäser P, Thomine S, Schroeder JI, et al. Phylogenetic relationships within cation transporter families of Arabidopsis[J]. Plant Physiol, 2001, 126(4): 1646-1667.
doi: 10.1104/pp.126.4.1646 pmid: 11500563 |
| [16] | Artimo P, Jonnalagedda M, Arnold K, et al. ExPASy: SIB bioinformatics resource portal[J]. Nucleic Acids Res, 2012, 40(Web Server issue): W597-W603. |
| [17] |
Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets[J]. Mol Biol Evol, 2016, 33(7): 1870-1874.
doi: 10.1093/molbev/msw054 pmid: 27004904 |
| [18] |
Chen CJ, Chen H, Zhang Y, et al. TBtools: an integrative toolkit developed for interactive analyses of big biological data[J]. Mol Plant, 2020, 13(8): 1194-1202.
doi: S1674-2052(20)30187-8 pmid: 32585190 |
| [19] | Bailey TL, Johnson J, Grant CE, et al. The MEME suite[J]. Nucleic Acids Res, 2015, 43(W1): W39-W49. |
| [20] |
Qin L, Han PP, Chen LY, et al. Genome-wide identification and expression analysis of NRAMP family genes in soybean(Glycine max L.)[J]. Front Plant Sci, 2017, 8: 1436.
doi: 10.3389/fpls.2017.01436 pmid: 28868061 |
| [21] | 杨猛. 水稻NRAMP家族基因在Mn和Cd转运中的功能研究[D]. 武汉: 华中农业大学, 2014. |
| Yang M. Functional analysis of rice NRAMP genes in Mn and Cd transport[D]. Wuhan: Huazhong Agricultural University, 2014. | |
| [22] | 陈娜. 沙棘修复尾矿坝的重金属迁移时间效应分析及预测[D]. 沈阳: 辽宁大学, 2021. |
| Chen N. Analysis and prediction of heavy metal migration time effect of sea buckthorn remediation tailings dam[D]. Shenyang: Liaoning University, 2021. | |
| [23] |
Wang PP, Moore BM, Panchy NL, et al. Factors influencing gene family size variation among related species in a plant family, Solanaceae[J]. Genome Biol Evol, 2018, 10(10): 2596-2613.
doi: 10.1093/gbe/evy193 pmid: 30239695 |
| [24] | Tian SK, Wang DD, Yang L, et al. A systematic review of 1-Deoxy-D-xylulose-5-phosphate synthase in terpenoid biosynthesis in plants[J]. Plant Growth Regul, 2022, 96(2): 221-235. |
| [25] | Mani A, Sankaranarayanan K. In silico analysis of natural resistance-associated macrophage protein(NRAMP)family of transporters in rice[J]. Protein J, 2018, 37(3): 237-247. |
| [26] |
Delsuc F, Brinkmann H, Philippe H. Phylogenomics and the reconstruction of the tree of life[J]. Nat Rev Genet, 2005, 6(5): 361-375.
doi: 10.1038/nrg1603 pmid: 15861208 |
| [27] | Chang JD, Gao WP, Wang P, et al. OsNRAMP5 is a major transporter for lead uptake in rice[J]. Environ Sci Technol, 2022, 56(23): 17481-17490. |
| [28] |
Gabaldón T, Koonin EV. Functional and evolutionary implications of gene orthology[J]. Nat Rev Genet, 2013, 14: 360-366.
doi: 10.1038/nrg3456 pmid: 23552219 |
| [29] | Tiwari M, Sharma D, Dwivedi S, et al. Expression in Arabidopsis and cellular localization reveal involvement of rice NRAMP, OsNRAMP1, in arsenic transport and tolerance[J]. Plant Cell Environ, 2014, 37(1): 140-152. |
| [30] | Nie YY, Li Y, Liu MH, et al. The nucleoporin NUP160 and NUP96 regulate nucleocytoplasmic export of mRNAs and participate in ethylene signaling and response in Arabidopsis[J]. Plant Cell Rep, 2023, 42(3): 549-559. |
| [31] | Martin RE, Marzol E, Estevez JM, et al. Ethylene signaling increases reactive oxygen species accumulation to drive root hair initiation in Arabidopsis[J]. Development, 2022, 149(13): dev200487. |
| [32] | Gao HY, Xia XY, An LJ. Critical roles of the activation of ethylene pathway genes mediated by DNA demethylation in Arabidopsis hyperhydricity[J]. Plant Genome, 2022, 15(2): e20202. |
| [33] | Guedes FRCM, Maia CF, Silva BRSD, et al. Exogenous 24-Epibrassinolide stimulates root protection, and leaf antioxidant enzymes in lead stressed rice plants: central roles to minimize Pb content and oxidative stress[J]. Environ Pollut, 2021, 280: 116992. |
| [34] |
Thao NP, Khan MI, Thu NBA, et al. Role of ethylene and its cross talk with other signaling molecules in plant responses to heavy metal stress[J]. Plant Physiol, 2015, 169(1): 73-84.
doi: 10.1104/pp.15.00663 pmid: 26246451 |
| [35] | Zou Y, Ding XC, Ye LL, et al. Molecular cloning and expression analysis of EIN2, EIN3/EIL, and EBF genes during papaya fruit development and ripening[J]. N Z J Crop Hortic Sci, 2021, 49(2/3): 151-167. |
| [1] | LOU Yin, GAO Hao-jun, WANG Xi, NIU Jing-ping, WANG Min, DU Wei-jun, YUE Ai-qin. Identification and Expression Pattern Analysis of GmHMGS Gene in Soybean [J]. Biotechnology Bulletin, 2024, 40(4): 110-121. |
| [2] | CHEN Chun-lin, LI Bai-xue, LI Jin-ling, DU Qing-jie, LI Meng, XIAO Huai-juan. Identification and Expression Analysis of Epidermal Patterning Factor (EPF) Genes in Cucumis melo [J]. Biotechnology Bulletin, 2024, 40(4): 130-138. |
| [3] | XIAO Ya-ru, JIA Ting-ting, LUO Dan, WU Zhe, LI Li-xia. Cloning and Expression Analysis of CsERF025L Transcription Factor in Cucumber [J]. Biotechnology Bulletin, 2024, 40(4): 159-166. |
| [4] | ZHONG Yun, LIN Chun, LIU Zheng-jie, DONG Chen-wen-hua, MAO Zi-chao, LI Xing-yu. Cloning and Prokaryotic Expression Analysis of Asparagus Saponin Synthesis Related Glycosyltransferase Genes [J]. Biotechnology Bulletin, 2024, 40(4): 255-263. |
| [5] | YANG Wei-cheng, SUN Yan, YANG Qian, WANG Zhuang-lin, MA Ju-hua, XUE Jin-ai, LI Run-zhi. Genome-wide Identification of the FAX family in Gossypium hirsutum and Functional Analysis of GhFAX1 [J]. Biotechnology Bulletin, 2024, 40(3): 155-169. |
| [6] | YANG Yan, HU Yang, LIU Ni-ru, YIN Lu, YANG Rui, WANG Peng-fei, MU Xiao-peng, ZHANG Shuai, CHENG Chun-zhen, ZHANG Jian-cheng. Cloning and Functional Analysis of MbbZIP43 Gene in ‘Hongmantang’ Red-flesh Apple [J]. Biotechnology Bulletin, 2024, 40(2): 146-159. |
| [7] | REN Yan-jing, ZHANG Lu-gang, ZHAO Meng-liang, LI Jiang, SHAO Deng-kui. cDNA Yeast Library Construction of Chinese Cabbage Seeds and Screening and Analysis of BrTTG1 Interacting Proteins [J]. Biotechnology Bulletin, 2024, 40(2): 223-232. |
| [8] | XIE Hong, ZHOU Li-ying, LI Shu-wen, WANG Meng-di, AI Ye, CHAO Yue-hui. Structural and Functional Analysis of MtCIM Gene in Medicago truncatula [J]. Biotechnology Bulletin, 2024, 40(1): 262-269. |
| [9] | TANG Wei-lin, KANG Qin, WANG Xia, SHEN Ming-yang, SUN Xin-jiang, WANG Ke, HOU Kai, WU Wei, XU Dong-bei. Cloning and Expression Pattern Analysis of Jasmonic Acid Receptor Gene McCOI1a in Mentha canadensis L. [J]. Biotechnology Bulletin, 2024, 40(1): 270-280. |
| [10] | JIANG Run-hai, JIANG Ran-ran, ZHU Cheng-qiang, HOU Xiu-li. Research Progress in Mechanisms of Microbial-enhanced Phytoremediation for Lead-contaminated Soil [J]. Biotechnology Bulletin, 2023, 39(8): 114-125. |
| [11] | SUN Ming-hui, WU Qiong, LIU Dan-dan, JIAO Xiao-yu, WANG Wen-jie. Cloning and Expression Analysis of CsTMFs Gene in Tea Plant [J]. Biotechnology Bulletin, 2023, 39(7): 151-159. |
| [12] | ZHAO Xue-ting, GAO Li-yan, WANG Jun-gang, SHEN Qing-qing, ZHANG Shu-zhen, LI Fu-sheng. Cloning and Expression of AP2/ERF Transcription Factor Gene ShERF3 in Sugarcane and Subcellular Localization of Its Encoded Protein [J]. Biotechnology Bulletin, 2023, 39(6): 208-216. |
| [13] | JIANG Qing-chun, DU Jie, WANG Jia-cheng, YU Zhi-he, WANG Yun, LIU Zhong-yu. Expression and Function Analysis of Transcription Factor PcMYB2 from Polygonum cuspidatum [J]. Biotechnology Bulletin, 2023, 39(5): 217-223. |
| [14] | YAO Zi-ting, CAO Xue-ying, XIAO Xue, LI Rui-fang, WEI Xiao-mei, ZOU Cheng-wu, ZHU Gui-ning. Screening of Reference Genes for RT-qPCR in Neoscytalidium dimidiatum [J]. Biotechnology Bulletin, 2023, 39(5): 92-102. |
| [15] | WANG Yi-qing, WANG Tao, WEI Chao-ling, DAI Hao-min, CAO Shi-xian, SUN Wei-jiang, ZENG Wen. Identification and Interaction Analysis of SMAS Gene Family in Tea Plant(Camellia sinensis) [J]. Biotechnology Bulletin, 2023, 39(4): 246-258. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||