








Biotechnology Bulletin ›› 2024, Vol. 40 ›› Issue (9): 282-290.doi: 10.13560/j.cnki.biotech.bull.1985.2024-0213
Previous Articles Next Articles
HOU Zhi-han( ), HAO Nan, LI Jia-qi, ZHAO Bin(
), HAO Nan, LI Jia-qi, ZHAO Bin( ), LIU Ying-chao(
), LIU Ying-chao( )
)
Received:2024-03-06
Online:2024-09-26
Published:2024-10-12
Contact:
ZHAO Bin, LIU Ying-chao
E-mail:15630858209@163.com;bdzhaobin@126.com;liuyingchao@hebau.edu.cn
HOU Zhi-han, HAO Nan, LI Jia-qi, ZHAO Bin, LIU Ying-chao. Roles of RNA m1A and m5C Methylation Modifications in the Fumonisin Biosynthesis of Fusarium verticillioides[J]. Biotechnology Bulletin, 2024, 40(9): 282-290.
| 引物名称 Primer name | 引物序列 Primer sequence(5'-3') |
|---|---|
| 18S ribosomal RNA-F 18S ribosomal RNA-R Fvnsun2-F Fvnsun2-R Fvnsun4-F Fvnsun4-R Fvalyref-F Fvalyref-R | GGCCGTTCTTAGTTGGTGGA TGCGGCCCAGAACATCTAAG AGACCTTCCGCAAGCTTCTC CGCTCTCTAAAGTCGGGGTC TGGGTGGTTTCGATCGTGTT TAAACAATGTAGCCGCCCGT AGGCCGTGCTATTGAAGTCC AAAGGGTTAGGAGGGACGGA |
Table 1 Primers used in the experiment
| 引物名称 Primer name | 引物序列 Primer sequence(5'-3') |
|---|---|
| 18S ribosomal RNA-F 18S ribosomal RNA-R Fvnsun2-F Fvnsun2-R Fvnsun4-F Fvnsun4-R Fvalyref-F Fvalyref-R | GGCCGTTCTTAGTTGGTGGA TGCGGCCCAGAACATCTAAG AGACCTTCCGCAAGCTTCTC CGCTCTCTAAAGTCGGGGTC TGGGTGGTTTCGATCGTGTT TAAACAATGTAGCCGCCCGT AGGCCGTGCTATTGAAGTCC AAAGGGTTAGGAGGGACGGA |
| 菌株编号 Strain number | 菌株来源 Strain source | FB1含量 Content of FB1/(mg·g-1) |
|---|---|---|
| 87 | 14-SD-94-6 | 145.20 |
| 64 | HK12-5 | 116.90 |
| 114 | 14-17-15 | 228.20 |
| 126 | 14-SD-91-13 | 116.80 |
| 81 | HN 22-2 | 112.30 |
| 104 | 25-4 | 89.55 |
| 50 | LZ-2-81 | 103.57 |
| 80 | NMG-1-2 | 92.44 |
| 84 | 124-3 | 64.73 |
| 86 | HN 24-2 | 99.39 |
| 63 | S-9-7 | 101.82 |
| 6 | M-I1-46 | 59.29 |
| 38 | M-10-48 | ND |
| 1 | LZ-2-77甘 | 2.18 |
| 49 | P-HB-T3-4 | 2.19 |
| 55 | NMG-3-3 | ND |
| 109 | M-3-20 | ND |
| 36 | 26-4 | ND |
| 18 | 14-SD-22-20 | ND |
| 37 22 | 14-SD-91-9 M-4-78 | ND ND |
| 40 139 | ZL-1-6 14-SD-91-9 | ND 1.81 |
| 33 | P-C1-32-1 | ND |
Table 2 Determination of FB1 content of F. verticillioides strain in different regions
| 菌株编号 Strain number | 菌株来源 Strain source | FB1含量 Content of FB1/(mg·g-1) |
|---|---|---|
| 87 | 14-SD-94-6 | 145.20 |
| 64 | HK12-5 | 116.90 |
| 114 | 14-17-15 | 228.20 |
| 126 | 14-SD-91-13 | 116.80 |
| 81 | HN 22-2 | 112.30 |
| 104 | 25-4 | 89.55 |
| 50 | LZ-2-81 | 103.57 |
| 80 | NMG-1-2 | 92.44 |
| 84 | 124-3 | 64.73 |
| 86 | HN 24-2 | 99.39 |
| 63 | S-9-7 | 101.82 |
| 6 | M-I1-46 | 59.29 |
| 38 | M-10-48 | ND |
| 1 | LZ-2-77甘 | 2.18 |
| 49 | P-HB-T3-4 | 2.19 |
| 55 | NMG-3-3 | ND |
| 109 | M-3-20 | ND |
| 36 | 26-4 | ND |
| 18 | 14-SD-22-20 | ND |
| 37 22 | 14-SD-91-9 M-4-78 | ND ND |
| 40 139 | ZL-1-6 14-SD-91-9 | ND 1.81 |
| 33 | P-C1-32-1 | ND |
| 核苷Nucleotide | 母离子Precursorion(m/z) | 子离子Production(m/z) | 保留时间Residence time/ ms | 去簇电压DP/V | 碰撞电压CE /V |
|---|---|---|---|---|---|
| Um | 259.3 | 113.1 | 0.94 | 17 | 14 |
| Gm | 298.2 | 152.1 | 1.10 | 8 | 16 |
| m1A | 282.1 | 150.1 | 0.72 | 36 | 30 |
| m6A | 282.2 | 150.2 | 2.40 | 39 | 20 |
| m5C | 258.3 | 126.1 | 0.68 | 24 | 18 |
| m7G | 298.1 | 166.1 | 0.76 | 20 | 18 |
Table 3 Retention time and mass spectrum parameters of 6 target nucleosides
| 核苷Nucleotide | 母离子Precursorion(m/z) | 子离子Production(m/z) | 保留时间Residence time/ ms | 去簇电压DP/V | 碰撞电压CE /V |
|---|---|---|---|---|---|
| Um | 259.3 | 113.1 | 0.94 | 17 | 14 |
| Gm | 298.2 | 152.1 | 1.10 | 8 | 16 |
| m1A | 282.1 | 150.1 | 0.72 | 36 | 30 |
| m6A | 282.2 | 150.2 | 2.40 | 39 | 20 |
| m5C | 258.3 | 126.1 | 0.68 | 24 | 18 |
| m7G | 298.1 | 166.1 | 0.76 | 20 | 18 |
| 核苷 Nucleotide | 检出限LOD /(μg·kg-1) | 基质效应 Matrix effect/% | 线性方程 Linearity equation | 决定系数R2 Determination coefficient | 平均回收率/相对标准偏差 Average recovery(%)/ RSD | |||
|---|---|---|---|---|---|---|---|---|
| 1 μg/kg | 20 μg/kg | 500 μg/kg | ||||||
| Um | 0.002 | 5.41 | y =4E+06x - 2964.3 | 0.9979 | 91.9/4.7 | 84.4/4.5 | 91.9/3 | |
| Gm | 0.001 | 2.27 | y =2E+06x - 5329.46 | 0.9999 | 97.6/6.6 | 86.2/7.8 | 109.7/4.9 | |
| m1A | 0.0009 | 2.16 | y = 7E+07x - 161526 | 0.9990 | 72.5/4.9 | 75.1/1.9 | 74.4/7.6 | |
| m6A | 0.0006 | 2.13 | y = 7E+07x - 30294 | 0.9981 | 88.5/7.0 | 78.7/5.9 | 116.5/1.4 | |
| m5C | 0.0003 | 2.51 | y = 3E+06x - 1896.4 | 0.9993 | 70.3/6.5 | 71.0/4.7 | 74.1/7.2 | |
| m7G | 0.0006 | 1.95 | y = 6E+07x - 141064 | 0.9995 | 74.0/8.9 | 72.1/7.7 | 70.9/9.5 | |
Table 4 Linear equation, determination coefficient, average recovery, and relative standard deviation of 6 nucleosides
| 核苷 Nucleotide | 检出限LOD /(μg·kg-1) | 基质效应 Matrix effect/% | 线性方程 Linearity equation | 决定系数R2 Determination coefficient | 平均回收率/相对标准偏差 Average recovery(%)/ RSD | |||
|---|---|---|---|---|---|---|---|---|
| 1 μg/kg | 20 μg/kg | 500 μg/kg | ||||||
| Um | 0.002 | 5.41 | y =4E+06x - 2964.3 | 0.9979 | 91.9/4.7 | 84.4/4.5 | 91.9/3 | |
| Gm | 0.001 | 2.27 | y =2E+06x - 5329.46 | 0.9999 | 97.6/6.6 | 86.2/7.8 | 109.7/4.9 | |
| m1A | 0.0009 | 2.16 | y = 7E+07x - 161526 | 0.9990 | 72.5/4.9 | 75.1/1.9 | 74.4/7.6 | |
| m6A | 0.0006 | 2.13 | y = 7E+07x - 30294 | 0.9981 | 88.5/7.0 | 78.7/5.9 | 116.5/1.4 | |
| m5C | 0.0003 | 2.51 | y = 3E+06x - 1896.4 | 0.9993 | 70.3/6.5 | 71.0/4.7 | 74.1/7.2 | |
| m7G | 0.0006 | 1.95 | y = 6E+07x - 141064 | 0.9995 | 74.0/8.9 | 72.1/7.7 | 70.9/9.5 | |
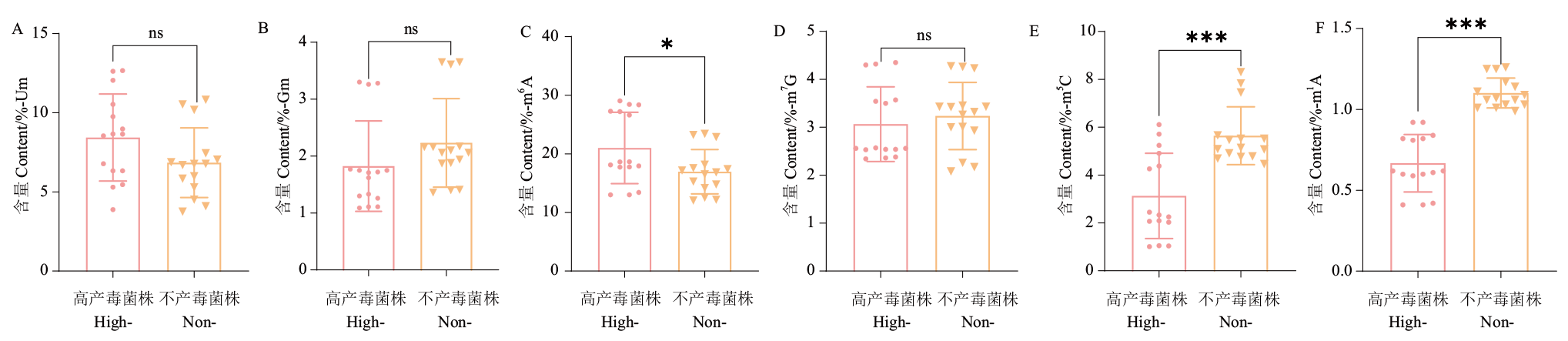
Fig. 2 Changes of RNA modification in toxicity-yielding differential strains A: Um modification content; B: Gm modification content; C: m6A modification content; D: m7G modification content; E: m5C modification content; F: m1A modification content. High-toxicity-producing strains abbreviated as High-. Non-toxicity-producing strains abbreviated as Non-. Data in the figure are mean ± SE. ns: No significant difference,* (P<0.05), ** (P<0.05), *** (P<0.001). The same below
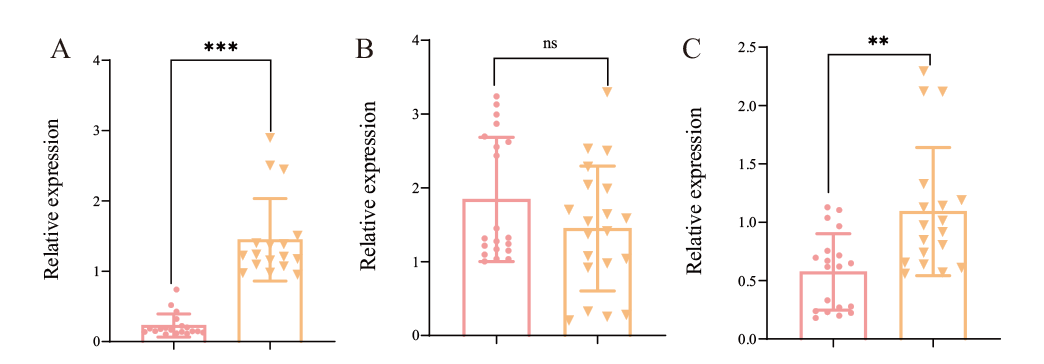
Fig. 4 Expression of m5C modification-related genes in different toxin-producing strains A: Expression of Fvalyref in the strains with different fumonisin contents; B-C: Expressions of Fvnsun2 and Fvnsun4 in the strains with different fumonisin content
| [1] | Deepa N, Achar PN, Sreenivasa MY. Current perspectives of biocontrol agents for management of Fusarium verticillioides and its fumonisin in cereals-a review[J]. J Fungi, 2021, 7(9): 776. |
| [2] |
Han SL, Wang MM, Ma ZY, et al. Fusarium diversity associated with diseased cereals in China, with an updated phylogenomic assessment of the genus[J]. Stud Mycol, 2023, 104: 87-148.
doi: 10.3114/sim.2022.104.02 pmid: 37351543 |
| [3] | Iqbal N, Czékus Z, Poór P, et al. Plant defence mechanisms against mycotoxin Fumonisin B1[J]. Chem Biol Interact, 2021, 343: 109494. |
| [4] | Carvajal-Moreno M. Mycotoxin challenges in maize production and possible control methods in the 21st century[J]. J Cereal Sci, 2022, 103: 103293. |
| [5] |
Winter G, Pereg L. A review on the relation between soil and mycotoxins: effect of aflatoxin on field, food and finance[J]. Eur J Soil Sci, 2019, 70(4): 882-897.
doi: 10.1111/ejss.12813 |
| [6] | Desmond J. A modest proposal: a response to the marketing challenges presented by the crisis confronting humanity in respect to the requirement to feed nine billion by 2050[J]. J Mark Manag, 2013, 29(13/14): 1631-1643. |
| [7] |
Dai YQ, Huang KL, Zhang BY, et al. Aflatoxin B1-induced epigenetic alterations: an overview[J]. Food Chem Toxicol, 2017, 109(Pt 1): 683-689.
doi: S0278-6915(17)30342-3 pmid: 28645871 |
| [8] | Teng PC, Liang YW, Yarmishyn AA, et al. RNA modifications and epigenetics in modulation of lung cancer and pulmonary diseases[J]. Int J Mol Sci, 2021, 22(19): 10592. |
| [9] | 马士清, 彭金英, 伊成器. RNA修饰检测技术[J]. 生命科学, 2018, 30(4): 440-446. |
| Ma SQ, Peng JY, Yi CQ. The detection methods of RNA modifications[J]. Chin Bull Life Sci, 2018, 30(4): 440-446. | |
| [10] | Yu HW, Raza SHA, Zhang WZ, et al. Research progress of m6A regulation during animal growth and development[J]. Mol Cell Probes, 2022, 65: 101851. |
| [11] | Ghazi T, Nagiah S, Chuturgoon AA. Fusaric acid induces hepatic global m6A RNA methylation and differential expression of m6A regulatory genes in vivo - a pilot study[J]. Epigenetics, 2022, 17(6): 695-703. |
| [12] |
Růžička K, Zhang M, Campilho A, et al. Identification of factors required for m6 A mRNA methylation in Arabidopsis reveals a role for the conserved E3 ubiquitin ligase HAKAI[J]. New Phytol, 2017, 215(1): 157-172.
doi: 10.1111/nph.14586 pmid: 28503769 |
| [13] |
Zou MJ, Mu Y, Chai X, et al. The critical function of the plastid rRNA methyltransferase, CMAL, in ribosome biogenesis and plant development[J]. Nucleic Acids Res, 2020, 48(6): 3195-3210.
doi: 10.1093/nar/gkaa129 pmid: 32095829 |
| [14] |
Zhang C, Zhang H, Zhu QQ, et al. Overexpression of global regulator LaeA increases secondary metabolite production in Monascus purpureus[J]. Appl Microbiol Biotechnol, 2020, 104(7): 3049-3060.
doi: 10.1007/s00253-020-10379-4 pmid: 32043189 |
| [15] | Wang B, Li XJ, Tabudravu J, et al. The chemical profile of activated secondary metabolites by overexpressing LaeA in Aspergillus niger[J]. Microbiol Res, 2021, 248: 126735. |
| [16] | Wang YS, Chen YC, Zhang J, et al. Overexpression of llm1 affects the synthesis of secondary metabolites of Aspergillus cristatus[J]. Microorganisms, 2022, 10(9): 1707. |
| [17] | Yadav PK, Rajasekharan R. The m6A methyltransferase Ime4 epitranscriptionally regulates triacylglycerol metabolism and vacuolar morphology in haploid yeast cells[J]. J Biol Chem, 2017, 292(33): 13727-13744. |
| [18] | Zhang YW, Wang YM, Fan JL, et al. Aspergillus fumigatus Elongator complex subunit 3 affects hyphal growth, adhesion and virulence through wobble uridine tRNA modification[J]. PLoS Pathog, 2022, 18(11): e1010976. |
| [19] | Ren ZY, Tang BZ, Xing JJ, et al. MTA1-mediated RNA m6 A modification regulates autophagy and is required for infection of the rice blast fungus[J]. New Phytol, 2022, 235(1): 247-262. |
| [20] |
Huang DY, Cui LQ, Sajid A, et al. The epigenetic mechanisms in Fusarium mycotoxins induced toxicities[J]. Food Chem Toxicol, 2019, 123: 595-601.
doi: S0278-6915(18)30798-1 pmid: 30599843 |
| [21] | 刘静. 拟轮枝镰孢ORPs家族主效蛋白鉴定及其功能解析[D]. 保定: 河北农业大学, 2023. |
| Liu J. Identification and functional analysis of major protein from ORPs family of Fusarium verticillioides[D]. Baoding: Hebei Agricultural University, 2023. | |
| [22] | Tian F, Lee SY, Woo SY, et al. Effect of plant-based compounds on the antifungal and antiaflatoxigenic efficiency of strobilurins against Aspergillus flavus[J]. J Hazard Mater, 2021, 415: 125663. |
| [23] | Bryła M, Roszko M, Szymczyk K, et al. Fumonisins in plant-origin food and fodder—a review[J]. Food Addit Contam Part A Chem Anal Control Expo Risk Assess, 2013, 30(9): 1626-1640. |
| [24] | Yang X, Gao J, Liu Q, et al. Co-occurrence of mycotoxins in maize and maize-derived food in China and estimation of dietary intake[J]. Food Addit Contam Part B Surveill, 2019, 12(2): 124-134. |
| [25] | 王燕, 董燕婕, 岳晖, 等. 山东省玉米真菌毒素污染状况调查及分析[J]. 粮油食品科技, 2016, 24(3): 69-73. |
| Wang Y, Dong YJ, Yue H, et al. Investigatin and analysis on mycotoxins contamination of maize in Shandong Province[J]. Sci Technol Cereals Oils Foods, 2016, 24(3): 69-73. | |
| [26] | Hu L, Liu HW, Yang J, et al. Free and hidden fumonisins in raw maize and maize-based products from China[J]. Food Addit Contam Part B Surveill, 2019, 12(2): 90-96. |
| [27] |
Covarelli L, Stifano S, Beccari G, et al. Characterization of Fusarium verticillioides strains isolated from maize in Italy: fumonisin production, pathogenicity and genetic variability[J]. Food Microbiol, 2012, 31(1): 17-24.
doi: 10.1016/j.fm.2012.02.002 pmid: 22475938 |
| [28] | Li LL, He ZQ, Shi Y, et al. Role of epigenetics in mycotoxin toxicity: a review[J]. Environ Toxicol Pharmacol, 2023, 100: 104154. |
| [29] | Yang C, Wu DD, Lin H, et al. Role of RNA modifications, especially m6A, in aflatoxin biosynthesis of Aspergillus flavus[J]. J Agric Food Chem, 2024, 72(1): 726-741. |
| [30] | Liang LK, Wang XY, Lan HE, et al. Comprehensive analysis of aflatoxin B1 biosynthesis in Aspergillus flavus via transcriptome-wide m6A methylome response to cycloleucine[J]. J Hazard Mater, 2024, 461: 132677. |
| [31] | Shafik AM, Zhou HQ, Lim J, et al. Dysregulated mitochondrial and cytosolic tRNA m1A methylation in Alzheimer's disease[J]. Hum Mol Genet, 2022, 31(10): 1673-1680. |
| [32] | Boo SH, Ha H, Kim YK. m1A and m6A modifications function cooperatively to facilitate rapid mRNA degradation[J]. Cell Rep, 2022, 40(10): 111317. |
| [33] | Qi ZY, Zhang C, Jian H, et al. N1-Methyladenosine modification of mRNA regulates neuronal gene expression and oxygen glucose deprivation/reoxygenation induction[J]. Cell Death Discov, 2023, 9(1): 159. |
| [34] | Trixl L, Lusser A. The dynamic RNA modification 5-methylcytosine and its emerging role as an epitranscriptomic mark[J]. Wiley Interdiscip Rev RNA, 2019, 10(1): e1510-e1527. |
| [35] | Zhao Y, Xing C, Peng HL. ALYREF(Aly/REF export factor): a potential biomarker for predicting cancer occurrence and therapeutic efficacy[J]. Life Sci, 2024, 338: 122372. |
| [36] |
Klec C, Knutsen E, Schwarzenbacher D, et al. ALYREF, a novel factor involved in breast carcinogenesis, acts through transcriptional and post-transcriptional mechanisms selectively regulating the short NEAT1 isoform[J]. Cell Mol Life Sci, 2022, 79(7): 391.
doi: 10.1007/s00018-022-04402-2 pmid: 35776213 |
| [37] | Wang N, Chen RX, Deng MH, et al. m5C-dependent cross-regulation between nuclear reader ALYREF and writer NSUN2 promotes urothelial bladder cancer malignancy through facilitating RABL6/TK1 mRNAs splicing and stabilization[J]. Cell Death Dis, 2023, 14(2): 139. |
| [38] | Berson A, Goodman LD, Sartoris AN, et al. Drosophila Ref1/ALYREF regulates transcription and toxicity associated with ALS/FTD disease etiologies[J]. Acta Neuropathol Commun, 2019, 7(1): 65. |
| [39] |
Kow RL, Black AH, Saxton AD, et al. Loss of aly/ALYREF suppresses toxicity in both tau and TDP-43 models of neurodegeneration[J]. GeroScience, 2022, 44(2): 747-761.
doi: 10.1007/s11357-022-00526-2 pmid: 35122183 |
| [40] | Sultana S, Bao WX, Shimizu M, et al. Frequency of three mutations in the fumonisin biosynthetic gene cluster ofFusarium fujikuroi that are predicted to block fumonisin production[J]. World Mycotoxin J, 2021, 14(1): 49-60. |
| [41] | Janevska S, Ferling I, Jojić K, et al. Self-protection against the sphingolipid biosynthesis inhibitor fumonisin B1 is conferred by a FUM cluster-encoded ceramide synthase[J]. mBio, 2020, 11(3): e00455-20. |
| [1] | WANG Chao-min, HE Mei-dan, WANG Wen-zhi, YUAN Qian-hua, ZHANG Shu-zhen, SHEN Lin-bo. Establishment and Application of Real-time PCR for Sugarcane Striate Virus [J]. Biotechnology Bulletin, 2024, 40(6): 126-133. |
| [2] | SANG Sen-hua. Detection of NK Cell Cytotoxicity: Real-time Dynamic Imaging-Based Analysis [J]. Biotechnology Bulletin, 2024, 40(4): 77-84. |
| [3] | YANG Wei-jie, YANG Zhou-lin, ZHU Hao-dong, WEI Yu, LIU Jun, LIU Xun. Study on the Properties and Functions of LchAD Protein, a Key Module of Lichenysin Synthase [J]. Biotechnology Bulletin, 2024, 40(3): 322-332. |
| [4] | YANG Wen-li, ZHU Li-li, CHEN Jian, CHEN Yan-xin, YAO Juan, JIANG Da-gang. Research Progress in the Reference Materials of Crop Pathogens in China [J]. Biotechnology Bulletin, 2024, 40(2): 31-37. |
| [5] | LIU Xing-yu, LI Jie, ZHU Long-jiao, LI Xiang-yang, XU Wen-tao. Aptamer of Pseudomonas aeruginosa: Acquiring and Application [J]. Biotechnology Bulletin, 2024, 40(1): 186-193. |
| [6] | ZHANG Xue-ping, LU Yu-qing, ZHANG Yue-qian, LI Xiao-juan. Advances in Plant Extracellular Vesicles and Analysis Techniques [J]. Biotechnology Bulletin, 2023, 39(5): 32-43. |
| [7] | ZHOU Xi-wen, CHENG Ke, ZHU Hong-liang. Research Progress in the Approaches to in vivo RNA Secondary Structure Profiling in Plants [J]. Biotechnology Bulletin, 2023, 39(2): 51-62. |
| [8] | GUO Wen-bo, LU Yang, SUI Li, ZHAO Yu, ZOU Xiao-wei, ZHANG Zheng-kun, LI Qi-yun. Preparation and Application of Polyclonal Antibodies Against Beauveria bassiana Mycovirus BbPmV-4 Coat Protein [J]. Biotechnology Bulletin, 2023, 39(10): 58-67. |
| [9] | LI Hui-jie, DONG Lian-hua, CHEN Gui-fang, LIU Si-yuan, YANG Jia-yi, YANG Jing-ya. Establishment of Droplet Digital PCR Assay for Quantitative Detection of Pseudomonas cocovenenans in Foods [J]. Biotechnology Bulletin, 2023, 39(1): 127-136. |
| [10] | CHEN Xiao-lin, LIU Yang-er, XU Wen-tao, GUO Ming-zhang, LIU Hui-lin. Application of Synthetic Biology Based Whole-cell Biosensor Technology in the Rapid Detection of Food Safety [J]. Biotechnology Bulletin, 2023, 39(1): 137-149. |
| [11] | HU Hai-yang, YING Wan-qin, HE Jun, LV Zhi-xian, XIE Xiao-ping, DENG Zhong-liang. Establishment and Application of ERA Real-time Fluorescence Method for Rapid Detection of Mycoplasma pneumoniae [J]. Biotechnology Bulletin, 2022, 38(9): 264-270. |
| [12] | GAO Wei-xin, HUANG Huo-qing, ZHAO Jing, ZHANG Xin, YANG Ning, YANG Hao-meng. Construction and Activity Verification of Ribonucleoprotein Complex for Gene Editing [J]. Biotechnology Bulletin, 2022, 38(8): 60-68. |
| [13] | LAN Xin-yue, LIU Ning-ning, ZHU Long-jiao, CHEN Xu, CHU Hua-shuo, LI Xiang-yang, DUAN Nuo, XU Wen-tao. Tetracycline Bivalent Aptamer Non-enzyme Label-free Sensor [J]. Biotechnology Bulletin, 2022, 38(3): 276-284. |
| [14] | LUO Xue-cong, AN Meng-nan, WU Yuan-hua, XIA Zi-hao. Applications of Recombinase Polymerase Amplification in Plant Virus Detection [J]. Biotechnology Bulletin, 2022, 38(2): 269-280. |
| [15] | KONG De-zhen, NIE Ying-bin, XU Hong-jun, CUI Feng-juan, MU Pei-yuan, TIAN Xiao-ming. Effects of Blend Seeding on the Yield,Purity and Yield Advantage of F1 in Three-line Hybrid Wheat [J]. Biotechnology Bulletin, 2022, 38(10): 132-139. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||