








Biotechnology Bulletin ›› 2025, Vol. 41 ›› Issue (1): 173-185.doi: 10.13560/j.cnki.biotech.bull.1985.2024-0479
Previous Articles Next Articles
WANG Zi-ao1,2,3,4( ), TIAN Rui5, CUI Yong-mei1,2,3,4, BAI Yi-xiong1,2,3,4, YAO Xiao-hua1,2,3,4, AN Li-kun1,2,3,4(
), TIAN Rui5, CUI Yong-mei1,2,3,4, BAI Yi-xiong1,2,3,4, YAO Xiao-hua1,2,3,4, AN Li-kun1,2,3,4( ), WU Kun-lun5(
), WU Kun-lun5( )
)
Received:2024-05-24
Online:2025-01-26
Published:2025-01-22
Contact:
AN Li-kun, WU Kun-lun
E-mail:13043832262@163.com;anlikun@163.com;wklqaaf@163.com
WANG Zi-ao, TIAN Rui, CUI Yong-mei, BAI Yi-xiong, YAO Xiao-hua, AN Li-kun, WU Kun-lun. Bioinformatics and Expression Pattern Analysis of HvnJAZ4 Gene in Hulless Barley[J]. Biotechnology Bulletin, 2025, 41(1): 173-185.
| 引物Primer | 引物序列Primer sequence(5'-3') |
|---|---|
| HvnJAZ4 | AGCCGGCTGGATGGAGAGGGA |
| TATTATATCGGATCAGATGT | |
| HvnJAZ4 promotor | CATGCTAACTGCTCCTAG |
| CCAGCCGGCTCGGGAA | |
| HvnJAZ4 qPCR | TCATCCTGTGACACTAGCCTCC |
| GACTTGCTGTTGTTGTCGTT | |
| HvnJAZ1 | ATGGATCTGCTGGA |
| TTACTGGGCCTT | |
| HvnNINJA1 | ATGGAGGATGGCCTTGA |
| TTAGTTTTGGGCCGAGGC | |
| HvnCOI1b | AGAAAGGGTGGGAGGGAGGAGGA |
| CCCAAGCGACGAGGGGCAATAAG | |
| 18SrRNA | ACGAGTCAGCCTTCGTCGT |
| GCGATCTCTGTGCATGATG | |
| HvnJAZ4-GFP | GCTCTAGAAGAAAGGGTGGGAGGGAGGAGGA |
| CGGGTACCCTCAGATGTGTAGTTTTGT |
Table 1 Primers’ names and sequences
| 引物Primer | 引物序列Primer sequence(5'-3') |
|---|---|
| HvnJAZ4 | AGCCGGCTGGATGGAGAGGGA |
| TATTATATCGGATCAGATGT | |
| HvnJAZ4 promotor | CATGCTAACTGCTCCTAG |
| CCAGCCGGCTCGGGAA | |
| HvnJAZ4 qPCR | TCATCCTGTGACACTAGCCTCC |
| GACTTGCTGTTGTTGTCGTT | |
| HvnJAZ1 | ATGGATCTGCTGGA |
| TTACTGGGCCTT | |
| HvnNINJA1 | ATGGAGGATGGCCTTGA |
| TTAGTTTTGGGCCGAGGC | |
| HvnCOI1b | AGAAAGGGTGGGAGGGAGGAGGA |
| CCCAAGCGACGAGGGGCAATAAG | |
| 18SrRNA | ACGAGTCAGCCTTCGTCGT |
| GCGATCTCTGTGCATGATG | |
| HvnJAZ4-GFP | GCTCTAGAAGAAAGGGTGGGAGGGAGGAGGA |
| CGGGTACCCTCAGATGTGTAGTTTTGT |
| 元件 Site name | 序列 Sequence | 功能 Function | 数量 Amouny |
|---|---|---|---|
| A-box | CCGTCC | 顺式作用调节元件Cis-acting regulatory element | 1 |
| ABRE | ACGTG | 参与脱落酸反应的顺式作用元件 Cis-acting element involved in the abscisic acid responsiveness | 5 |
| ARE | AAACCA | 对厌氧诱导调节顺式作用元件 Cis-acting regulatory element essential for the anaerobic induction | 3 |
| AuxRR-core | GGTCCAT | 参与生长素反应的顺式作用调节元件 Cis-acting regulatory element involved in auxin responsiveness | 1 |
| Box 4 | ATTAAT | 参与光响应的保守DNA模块的一部分 Part of a conserved DNA module involved in light responsiveness | 1 |
| CAAT-box | CCAAT; CAAAT | 启动子和增强子区域中常见的顺式作用元件 Common Cis-acting element in promoter and enhancer regions | 10 |
| CAT-box | GCCACT | 与分生组织表达相关的顺式作用调控元件 Cis-acting regulatory element related to meristem expression | 3 |
| CGTCA-motif | CGTCA | 参与MeJA反应性的顺式作用调节元件 Cis-acting regulatory element involved in the MeJA-responsiveness | 2 |
| G-Box | CACGTC | 参与光响应的顺式作用调节元件 Cis-acting regulatory element involved in light responsiveness | 2 |
| G-box | CACGTC; CACGAC; GCCACGTGGA | 参与光响应的顺式作用调节元件 Cis-acting regulatory element involved in light responsiveness | 5 |
| GC-motif | CCCCCG | 参与缺氧特异性诱导的增强子样元件 Enhancer-like element involved in anoxic specific inducibility | 2 |
| I-box | CCATATCCAAT; CGATAAGGCG | 光响应元件Part of a light responsive element | 2 |
| LTR | CCGAAA | 参与低温响应的顺式作用元件 Cis-acting element involved in low-temperature responsiveness | 1 |
| MBS | CAACTG | MYB结合位点参与干旱诱导MYB binding site involved in drought-inducibility | 1 |
| O2-site | GATGATGTGG | 参与醇溶蛋白代谢调节的顺式作用调节元件 Cis-acting regulatory element involved in zein metabolism regulation | 1 |
| TATA-box | TATATA; ATATAT; TATA; ACAAAA; TATAA; TATATTTATATTT | 启动子核心元件Core promoter element around -30 of transcription start | 46 |
| TATC-box | TATCCCA | 参与赤霉素反应的顺式作用元件 Cis-acting element involved in gibberellin-responsiveness | 1 |
| TC-rich repeats | GTTTTCTTAC | 参与防御和压力反应的顺式作用元件 Cis-acting element involved in defense and stress responsiveness | 1 |
| TCA-element | CCATCTTTTT; TCAGAAGAGG | 参与水杨酸反应性的顺式作用元件 Cis-acting element involved in salicylic acid responsiveness | 2 |
| TCCC-motif | TCTCCCT | 光响应元件Part of a light responsive element | 3 |
| TGACG-motif | TGACG | 参与MeJA反应性的顺式作用调节元件 Cis-acting regulatory element involved in the MeJA-responsiveness | 2 |
| Unnamed_5 | TGTAATAATATATTTATATT | SEF1因子结合位点SEF1 factor binding site | 1 |
Table 2 Prediction of elements in the promoter region of HvnJAZ4 gene in hulless barely
| 元件 Site name | 序列 Sequence | 功能 Function | 数量 Amouny |
|---|---|---|---|
| A-box | CCGTCC | 顺式作用调节元件Cis-acting regulatory element | 1 |
| ABRE | ACGTG | 参与脱落酸反应的顺式作用元件 Cis-acting element involved in the abscisic acid responsiveness | 5 |
| ARE | AAACCA | 对厌氧诱导调节顺式作用元件 Cis-acting regulatory element essential for the anaerobic induction | 3 |
| AuxRR-core | GGTCCAT | 参与生长素反应的顺式作用调节元件 Cis-acting regulatory element involved in auxin responsiveness | 1 |
| Box 4 | ATTAAT | 参与光响应的保守DNA模块的一部分 Part of a conserved DNA module involved in light responsiveness | 1 |
| CAAT-box | CCAAT; CAAAT | 启动子和增强子区域中常见的顺式作用元件 Common Cis-acting element in promoter and enhancer regions | 10 |
| CAT-box | GCCACT | 与分生组织表达相关的顺式作用调控元件 Cis-acting regulatory element related to meristem expression | 3 |
| CGTCA-motif | CGTCA | 参与MeJA反应性的顺式作用调节元件 Cis-acting regulatory element involved in the MeJA-responsiveness | 2 |
| G-Box | CACGTC | 参与光响应的顺式作用调节元件 Cis-acting regulatory element involved in light responsiveness | 2 |
| G-box | CACGTC; CACGAC; GCCACGTGGA | 参与光响应的顺式作用调节元件 Cis-acting regulatory element involved in light responsiveness | 5 |
| GC-motif | CCCCCG | 参与缺氧特异性诱导的增强子样元件 Enhancer-like element involved in anoxic specific inducibility | 2 |
| I-box | CCATATCCAAT; CGATAAGGCG | 光响应元件Part of a light responsive element | 2 |
| LTR | CCGAAA | 参与低温响应的顺式作用元件 Cis-acting element involved in low-temperature responsiveness | 1 |
| MBS | CAACTG | MYB结合位点参与干旱诱导MYB binding site involved in drought-inducibility | 1 |
| O2-site | GATGATGTGG | 参与醇溶蛋白代谢调节的顺式作用调节元件 Cis-acting regulatory element involved in zein metabolism regulation | 1 |
| TATA-box | TATATA; ATATAT; TATA; ACAAAA; TATAA; TATATTTATATTT | 启动子核心元件Core promoter element around -30 of transcription start | 46 |
| TATC-box | TATCCCA | 参与赤霉素反应的顺式作用元件 Cis-acting element involved in gibberellin-responsiveness | 1 |
| TC-rich repeats | GTTTTCTTAC | 参与防御和压力反应的顺式作用元件 Cis-acting element involved in defense and stress responsiveness | 1 |
| TCA-element | CCATCTTTTT; TCAGAAGAGG | 参与水杨酸反应性的顺式作用元件 Cis-acting element involved in salicylic acid responsiveness | 2 |
| TCCC-motif | TCTCCCT | 光响应元件Part of a light responsive element | 3 |
| TGACG-motif | TGACG | 参与MeJA反应性的顺式作用调节元件 Cis-acting regulatory element involved in the MeJA-responsiveness | 2 |
| Unnamed_5 | TGTAATAATATATTTATATT | SEF1因子结合位点SEF1 factor binding site | 1 |
| 理化性质Physicochemical property | HvnJAZ4 | 理化性质Physicochemical property | HvnJAZ4 | |
|---|---|---|---|---|
| 分子重量Molecular weight/Da | 44 055.39 | α 螺旋比例Proportions of α helix/% | 23.50 | |
| 总原子数Total number of atoms | 6 124 | 延伸链比例Proportions of extended strand/% | 12.47 | |
| 分子式Formula | C1908H3030N568O603S12 | β-折叠比例Proportions of β turn/% | 2.64 | |
| 亲水系数GRAVY | -0.497 | 无规则卷曲比例Proportions of random coil/% | 61.39 | |
| 理论等电点Theoretical pI | 10.0 | 亚细胞定位预测Prediction of subcellular localization | 细胞核Nucleus | |
| 不稳定指数Instability index(II) | 71.38 | 信号肽Signal peptide | 无No | |
| 跨膜结构Transmembrane structures | 无No | 磷酸化位点Phosphorylation site | 丝氨酸43个 | |
| 糖基化位点Glycosylation site 脂溶指数Aliphatic index | 无No 57.03 | 苏氨酸11个 酪氨酸2个 |
Table 3 Physical and chemical properties of HvnJAZ4 protein in hulless barely
| 理化性质Physicochemical property | HvnJAZ4 | 理化性质Physicochemical property | HvnJAZ4 | |
|---|---|---|---|---|
| 分子重量Molecular weight/Da | 44 055.39 | α 螺旋比例Proportions of α helix/% | 23.50 | |
| 总原子数Total number of atoms | 6 124 | 延伸链比例Proportions of extended strand/% | 12.47 | |
| 分子式Formula | C1908H3030N568O603S12 | β-折叠比例Proportions of β turn/% | 2.64 | |
| 亲水系数GRAVY | -0.497 | 无规则卷曲比例Proportions of random coil/% | 61.39 | |
| 理论等电点Theoretical pI | 10.0 | 亚细胞定位预测Prediction of subcellular localization | 细胞核Nucleus | |
| 不稳定指数Instability index(II) | 71.38 | 信号肽Signal peptide | 无No | |
| 跨膜结构Transmembrane structures | 无No | 磷酸化位点Phosphorylation site | 丝氨酸43个 | |
| 糖基化位点Glycosylation site 脂溶指数Aliphatic index | 无No 57.03 | 苏氨酸11个 酪氨酸2个 |

Fig. 1 Protein sequence analysis of HvnJAZ4 in hulless barely *: Key amino acid sites for the hydrophobic interaction between HvnJAZ4·jas and HvnCOI1b. ●: Key amino acid sites for the hydrogen bond interaction between HvnJAZ4·jas and HvnCOI1b
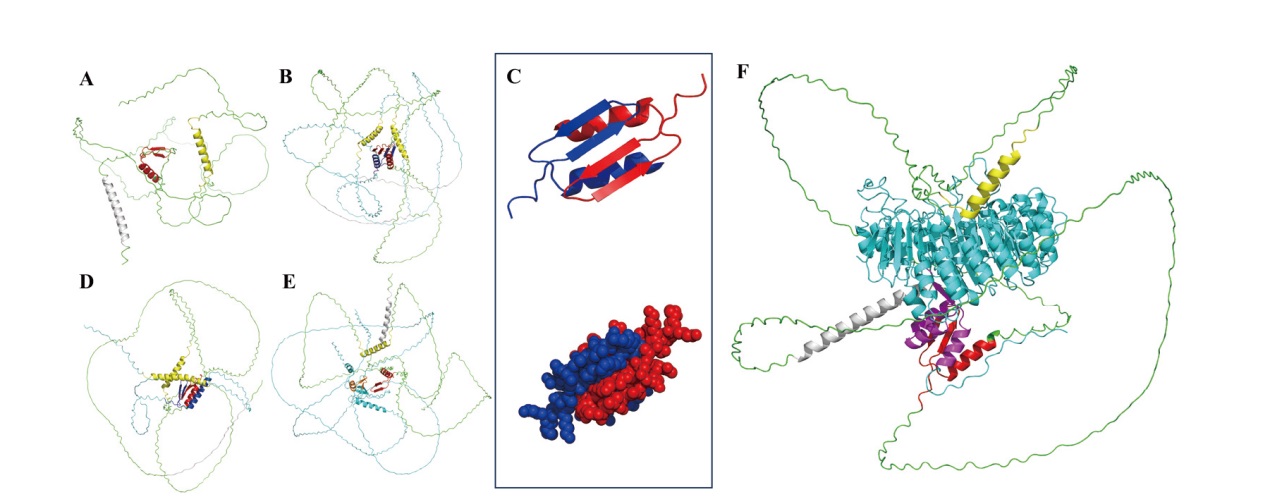
Fig. 3 Interaction model prediction of HvnJAZ4 with HvnJAZ4, HvnJAZ1, HvnNINJA1, and HvnCOI1b A: HvnJAZ4. B: Interaction model of HvnJAZ4 and HvnJAZ4. C: Interaction model of HvnJAZ4 ZIM domain. D: Interaction model of HvnJAZ4 and HvnJAZ1. E: Interaction model of HvnJAZ4 and HvnNINJA1. F: Interaction model of HvnJAZ4 and HvnCOI1b, Jas domain(yellow), ZIM domain(red, bule), NT domain(gray), HvnJAZ4(green), HvnJAZ1(cyans)HvnNINJA1(cyans), HvnNINJA1 TDBD domain(orange), HvnCOI1b(cyans), HvnCOI1b F-box domain(purple)

Fig. 4 RMSD and RMSF analysis of the HvnCOI1b-HvnJAZ4·jas complex and free protein HvnCOI1b during the 30 ns simulation A: RMSD analysis; B: RMSF analysis

Fig. 5 Decomposition of the binding energy on a per-residue basis for HvnCOI1b and HvnJAZ4·jas during the 30 ns simulation A: Decomposition of the binding energy on a per-residue basis for HvnCOI1b in the HvnCOI1b-HvnJAZ4 complex. B: Decomposition of the binding energy on a per-residue basis for HvnJAZ4·jas in the HvnCOI1b-HvnJAZ9 complex

Fig. 6 Binding model of HvnCOI1b-HvnJAZ4·jas complex A: Interaction model of JA-Ile, HvnCOI1b and HvnJAZ4·Jas, JA-Ile(yellow), HvnCOI1b(green), and HvnJAZ4·Jas(rose). B: Key amino acid sites analysis of the interaction of JA-Ile, HvnCOI1b, and HvnJAZ4·jas interaction

Fig. 7 Expression pattern of HvnJAZ4 in hulless barley A: Expression of HvnJAZ4 in different tissues. B: Expression of HvnJAZ4 under low temperatures treatments. C-H: Expression of HvnJAZ4 under MeJA, ABA, SA, GA, NAA and 6-BA treatments. Different lowercase letters indicate significant differences(P<0.05)

Fig. 8 Subcellular localization of HvnJAZ4 protein in hulless barley A: The fluorescence field of 35:HvnJAZ4-GFP. B: The chlorophyll autofluorescence field of 35:HvnJAZ4-GFP. C: The bright field of 35:HvnJAZ4-GFP. D: The merged field of 35:HvnJAZ4-GFP. E: The fluorescence field of 35:GFP. F: The chlorophyll autofluorescence field of 35:GFP. G: The bright field of 35:GFP. H: The merged field of 35:GFP
| [1] | 安立昆, 姚有华, 姚晓华, 等. 青稞耐低氮相关类甜蛋白基因HvTOND1克隆和亚细胞定位研究[J]. 西北农业学报, 2021, 30(8): 1157-1166. |
| An LK, Yao YH, Yao XH, et al. Cloning and subcellular localization of related thaumatin-like protein gene HvTOND1 tolerant to low nitrogen in hulless barley[J]. Acta Agric Boreali Occidentalis Sin, 2021, 30(8): 1157-1166. | |
| [2] | 任晴雯, 安立昆, 姚有华, 等. 青稞HvnPHO1;2基因克隆、亚细胞定位和表达模式分析[J]. 西北农业学报, 2021, 30(10): 1461-1472. |
| Ren QW, An LK, Yao YH, et al. Cloning,subcellular localization and expression analysis of phosphate transporter gene HvPHO1;2 in hulless barely[J]. Acta Agriculturae Boreali - occidentalis Sinica, 2021, 30(10): 1461-1472. | |
| [3] | 颜昌兰. 青稞品种稳定性、适应性及主要农艺性状的评价与分析[D]. 杨凌: 西北农林科技大学, 2015. |
| Yan CL. Evaluation and analysis of stability, adaptability and main agronomic characters of highland barley varieties[D]. Yangling: Northwest A&F University, 2015. | |
| [4] | 弓开元. 青藏高原青稞产量和光温生产潜力对气候变化的响应[D]. 杨凌: 西北农林科技大学, 2020. |
| Gong KY. Response of highland barley yield and light and temperature production potential to climate change in Qinghai-Tibet Plateau[D]. Yangling: Northwest A&F University, 2020. | |
| [5] | 张银乐, 张文静, 杨洋, 等. 低温对青稞种子萌发及幼苗生长的影响[J]. 安徽农学通报, 2018, 24(16): 28-31. |
| Zhang YL, Zhang WJ, Yang Y, et al. Effects of low temperature on seed germination and seedling growth of highland barley[J]. Anhui Agric Sci Bull, 2018, 24(16): 28-31. | |
| [6] | 王玉林, 徐齐君, 原红军, 等. 西藏青稞品种喜马拉雅8号低温处理的SSH-cDNA文库构建及分析[J]. 麦类作物学报, 2017, 37(8): 1025-1030. |
| Wang YL, Xu QJ, Yuan HJ, et al. Construction and analyses of SSH-cDNA library of Tibetan hulless barley(Hordeum vulgare l. var. nudum HK.f.)ximalaya 8 under low temperature treatment[J]. J Triticeae Crops, 2017, 37(8): 1025-1030. | |
| [7] | Yan YX, Christensen S, Isakeit T, et al. Disruption of OPR7 and OPR8 reveals the versatile functions of jasmonic acid in maize development and defense[J]. Plant Cell, 2012, 24(4): 1420-1436. |
| [8] | An LK, Ahmad RM, Ren H, et al. Jasmonate signal receptor gene family ZmCOIs restore male fertility and defense response of Arabidopsis mutant coi1-1[J]. Plant Growth Regul, 2019, 38(2): 479-493. |
| [9] | Chini A, Fonseca S, Fernández G, et al. The JAZ family of repressors is the missing link in jasmonate signalling[J]. Nature, 2007, 448(7154): 666-671. |
| [10] | Chung HS, Cooke TF, Depew CL, et al. Alternative splicing expands the repertoire of dominant JAZ repressors of jasmonate signaling[J]. Plant J, 2010, 63(4): 613-622. |
| [11] | 孙程, 周晓今, 陈茹梅, 等. 植物JAZ蛋白的功能概述[J]. 生物技术通报, 2014(6): 1-8. |
| Sun C, Zhou XJ, Chen RM, et al. Comprehensive overview of JAZ proteins in plants[J]. Biotechnol Bull, 2014(6): 1-8. | |
| [12] | Thines B, Katsir L, Melotto M, et al. JAZ repressor proteins are targets of the SCFCOI1 complex during jasmonate signalling[J]. Nature, 2007, 448: 661-665. |
| [13] |
Oblessuc PR, Obulareddy N, DeMott L, et al. JAZ4 is involved in plant defense, growth, and development in Arabidopsis[J]. Plant J, 2020, 101(2): 371-383.
doi: 10.1111/tpj.14548 |
| [14] |
晏胜伟, 孙程, 周晓今, 等. 玉米JAZ家族基因ZmJAZ4的克隆及功能分析[J]. 生物技术通报, 2015(3): 96-101.
doi: 10.13560/j.cnki.biotech.bull.1985.2015.04.013 |
| Yan SW, Sun C, Zhou XJ, et al. Cloning and characterization analysis of ZmJAZ4, a JAZ family gene in maize[J]. Biotechnol Bull, 2015,(3): 96-101. | |
| [15] | 王世通. 基于PhJAZ4转基因的矮牵牛新种质创制与评价[D]. 武汉: 华中农业大学, 2023. |
| Wang ST. Creation and evaluation of new petunia germplasm based on PhJAZ4 transgenic[D]. Wuhan: Huazhong Agricultural University, 2023. | |
| [16] | 杨娆. 丹参SmJAZ4调控酚酸类物质合成的分子机制[D]. 西安: 陕西师范大学, 2022. |
| Yang R. Molecular mechanism of the regulation of phenolic acid synthesis by Salvia miltiorrhiza SmJAZ4[D]. Xi'an: Shaanxi Normal University, 2022. | |
| [17] | Miccono MLA, Yang HW, DeMott L, et al. Review: Losing JAZ4 for growth and defense[J]. Plant Sci, 2023, 335: 111816. |
| [18] | Zhang MF, Luo X, He W, et al. OsJAZ4 fine-tunes rice blast resistance and yield traits[J]. Plants, 2024, 13(3): 348. |
| [19] | DeMott L, Oblessuc PR, Pierce A, et al. Spatiotemporal regulation of JAZ4 expression and splicing contribute to ethylene- and auxin-mediated responses in Arabidopsis roots[J]. Plant J, 2021, 108(5): 1266-1282. |
| [20] | Sheard LB, Tan X, Mao HB, et al. Jasmonate perception by inositol-phosphate-potentiated COI1-JAZ co-receptor[J]. Nature, 2010, 468(7322): 400-405. |
| [21] | Abramson J, Adler J, Dunger J, et al. Accurate structure prediction of biomolecular interactions with AlphaFold 3[J]. Nature, 2024, 630(8016): 493-500. |
| [22] |
Sanner MF. Python: a programming language for software integration and development[J]. J Mol Graph Model, 1999, 17(1): 57-61.
pmid: 10660911 |
| [23] |
Morris GM, Huey R, Lindstrom W, et al. AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility[J]. J Comput Chem, 2009, 30(16): 2785-2791.
doi: 10.1002/jcc.21256 pmid: 19399780 |
| [24] |
Trott O, Olson AJ. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading[J]. J Comput Chem, 2010, 31(2): 455-461.
doi: 10.1002/jcc.21334 pmid: 19499576 |
| [25] |
Pierce LCT, Salomon-Ferrer R, Augusto F de Oliveira C, et al. Routine access to millisecond time scale events with accelerated molecular dynamics[J]. J Chem Theory Comput, 2012, 8(9): 2997-3002.
pmid: 22984356 |
| [26] |
Götz AW, Williamson MJ, Xu D, et al. Routine microsecond molecular dynamics simulations with AMBER on GPUs. 1. generalized born[J]. J Chem Theory Comput, 2012, 8(5): 1542-1555.
pmid: 22582031 |
| [27] |
Salomon-Ferrer R, Götz AW, Poole D, et al. Routine microsecond molecular dynamics simulations with AMBER on GPUs. 2. explicit solvent particle mesh Ewald[J]. J Chem Theory Comput, 2013, 9(9): 3878-3888.
doi: 10.1021/ct400314y pmid: 26592383 |
| [28] |
Wang JM, Wolf RM, Caldwell JW, et al. Development and testing of a general amber force field[J]. J Comput Chem, 2004, 25(9): 1157-1174.
doi: 10.1002/jcc.20035 pmid: 15116359 |
| [29] |
Wang JM, Wang W, Kollman PA, et al. Automatic atom type and bond type perception in molecular mechanical calculations[J]. J Mol Graph Model, 2006, 25(2): 247-260.
pmid: 16458552 |
| [30] |
Sousa da Silva AW, Vranken WF. ACPYPE-AnteChamber PYthon parser interface[J]. BMC Res Notes, 2012, 5: 367.
doi: 10.1186/1756-0500-5-367 pmid: 22824207 |
| [31] | 魏昕, 刘雨恒, 刘宇阳, 等. 植物JAZ蛋白家族研究进展[J]. 植物生理学报, 2021, 57(5): 1039-1046. |
| Wei X, Liu YH, Liu YY, et al. Advances of JAZ family in plants[J]. China Ind Econ, 2021, 57(5): 1039-1046. | |
| [32] | Fu J, Wu H, Ma SQ, et al. OsJAZ1 attenuates drought resistance by regulating JA and ABA signaling in rice[J]. Front Plant Sci, 2017, 8: 2108. |
| [33] | 黄文峰, 王立丰, 田维敏. 茉莉酸反应基因转录抑制因子JAZ蛋白家族研究进展[J]. 热带作物学报, 2009, 30(9): 1383-1387. |
| Huang WF, Wang LF, Tian WM. Advances in JAZ protein family mediating JA signal pathway[J]. Chin J Trop Crops, 2009, 30(9): 1383-1387. | |
| [34] | 闫会转. JAZ2和JAZ7调控茉莉酸介导的转录与代谢重编程的机理研究[D]. 杭州: 浙江大学, 2014. |
| Yan HZ. Mechanism of JAZ2 and JAZ7 in regulating jasmonic acid mediated transcription and metabolic reprogramming[D]. Hangzhou: Zhejiang University, 2014. | |
| [35] | 安立昆. 玉米茉莉酸信号受体基因家族ZmCOIs功能研究[D]. 南京: 南京农业大学, 2018. |
| An LK. Study on the functional of maize jasmonic acid signal receptor gene family ZmCOIs[D]. Nanjing: Nanjing Agricultural University, 2018. | |
| [36] |
Saito R, Hayashi K, Nomoto H, et al. Extended JAZ degron sequence for plant hormone binding in jasmonate co receptor of tomato S1COI1 S1JAZ[J]. Sci Rep, 2021, 11(1): 13612.
doi: 10.1038/s41598-021-93067-1 pmid: 34193940 |
| [1] | KONG Qing-yang, ZHANG Xiao-long, LI Na, ZHANG Chen-jie, ZHANG Xue-yun, YU Chao, ZHANG Qi-xiang, LUO Le. Identification and Expression Analysis of GRAS Transcription Factor Family in Rosa persica [J]. Biotechnology Bulletin, 2025, 41(1): 210-220. |
| [2] | SONG Bing-fang, LIU Ning, CHENG Xin-yan, XU Xiao-bin, TIAN Wen-mao, GAO Yue, BI Yang, WANG Yi. Identification of Potato G6PDH Gene Family and Its Expression Analysis in Damaged Tubers [J]. Biotechnology Bulletin, 2024, 40(9): 104-112. |
| [3] | WU Hui-qin, WANG Yan-hong, LIU Han, SI Zheng, LIU Xue-qing, WANG Jing, YANG Yi, CHENG Yan. Identification and Expression Analysis of UGT Gene Family in Pepper [J]. Biotechnology Bulletin, 2024, 40(9): 198-211. |
| [4] | WU Juan, WU Xiao-juan, WANG Pei-jie, XIE Rui, NIE Hu-shuai, LI Nan, MA Yan-hong. Screening and Expression Analysis of ERF Gene Related to Anthocyanin Synthesis in Colored Potato [J]. Biotechnology Bulletin, 2024, 40(9): 82-91. |
| [5] | WU Shuai, XIN Yan-ni, MAI Chun-hai, MU Xiao-ya, WANG Min, YUE Ai-qin, ZHAO Jin-zhong, WU Shen-jie, DU Wei-jun, WANG Li-xiang. Genome-wide Identification and Stress Response Analysis of Soybean GS Gene Family [J]. Biotechnology Bulletin, 2024, 40(8): 63-73. |
| [6] | LIU Rong, TIAN Min-yu, LI Guang-ze, TAN Cheng-fang, RUAN Ying, LIU Chun-lin. Identification and Induced-expression Analysis of REVEILLE Family in Brassica napus L. [J]. Biotechnology Bulletin, 2024, 40(6): 161-171. |
| [7] | LI Jia-xin, LI Hong-yan, LIU Li-e, ZHANG Tian, ZHOU Wu. Identification and Expression Analysis of the NRAMP Family in Seabuckthorn Under Lead Stress [J]. Biotechnology Bulletin, 2024, 40(5): 191-202. |
| [8] | ZHONG Yun, LIN Chun, LIU Zheng-jie, DONG Chen-wen-hua, MAO Zi-chao, LI Xing-yu. Cloning and Prokaryotic Expression Analysis of Asparagus Saponin Synthesis Related Glycosyltransferase Genes [J]. Biotechnology Bulletin, 2024, 40(4): 255-263. |
| [9] | HAO Nan, GENG Shan, ZHAO Yu-wei, HOU Zhi-han, ZHAO Bin, LIU Ying-chao. Cloning and Expression Analysis of FvALT from Fusarium verticillioides [J]. Biotechnology Bulletin, 2024, 40(12): 256-263. |
| [10] | YANG Chong, CHENG Sha-sha, AI Chang-feng, ZHAO Xuan, LIU Meng-jun. Identification of ABF/AREB Gene Family and Their Expression Analysis in Jujube Fruit [J]. Biotechnology Bulletin, 2024, 40(11): 184-191. |
| [11] | SHI Jing-hui, CHEN Wen-hui, LU Kun, ZHENG Ting-ting, REN Zhi-yuan, BAO Guo-qing, WANG Min, LUO Jian-mei. Site-directed Saturation Mutagenesis to Improve the Catalytic Performance of 11α-hydroxylase from Aspergillus ochraceus [J]. Biotechnology Bulletin, 2024, 40(1): 322-331. |
| [12] | WANG Yi-qing, WANG Tao, WEI Chao-ling, DAI Hao-min, CAO Shi-xian, SUN Wei-jiang, ZENG Wen. Identification and Interaction Analysis of SMAS Gene Family in Tea Plant(Camellia sinensis) [J]. Biotechnology Bulletin, 2023, 39(4): 246-258. |
| [13] | PING Huai-lei, GUO Xue, YU Xiao, SONG Jing, DU Chun, WANG Juan, ZHANG Huai-bi. Cloning and Expression of PdANS in Paeonia delavayi and Correlation with Anthocyanin Content [J]. Biotechnology Bulletin, 2023, 39(3): 206-217. |
| [14] | GUO Zhi-hao, JIN Ze-xin, LIU Qi, GAO Li. Bioinformatics Analysis, Subcellular Localization and Toxicity Verification of Effector g11335 in Tilletia contraversa Kühn [J]. Biotechnology Bulletin, 2022, 38(8): 110-117. |
| [15] | YU Qiu-lin, MA Jing-yi, ZHAO Pan, SUN Peng-fang, HE Yu-mei, LIU Shi-biao, GUO Hui-hong. Cloning and Functional Analysis of Gynostemma pentaphyllum GpMIR156a and GpMIR166b [J]. Biotechnology Bulletin, 2022, 38(7): 186-193. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||