








Biotechnology Bulletin ›› 2024, Vol. 40 ›› Issue (8): 142-151.doi: 10.13560/j.cnki.biotech.bull.1985.2024-0257
Previous Articles Next Articles
ZHOU Lin1( ), HUANG Shun-man1, SU Wen-kun1, YAO Xiang2, QU Yan1(
), HUANG Shun-man1, SU Wen-kun1, YAO Xiang2, QU Yan1( )
)
Received:2024-03-16
Online:2024-08-26
Published:2024-07-31
Contact:
QU Yan
E-mail:1617574776@qq.com;quyan@swfu.edu.cn
ZHOU Lin, HUANG Shun-man, SU Wen-kun, YAO Xiang, QU Yan. Identification of the bHLH Gene Family and Selection of Genes Related to Color Formation in Camellia reticulata[J]. Biotechnology Bulletin, 2024, 40(8): 142-151.
| 基因Gene | 上游引物Forward primer(5'-3') | 下游引物Reverse primer(5'-3') |
|---|---|---|
| CrbHLH8 | GCAGTTGTGCCCTCAACAAG | CACTCACCTCCATTGGCTGT |
| CrbHLH13 | CGGCTGAATGCTCGTTGATG | CTGAAATTGGCGGTGTTGGG |
| CrbHLH59 | ACCCCATTGAATTCCCCACC | TACAAATCCAGGATCCGCGG |
| CrbHLH61 | GCAGTTGTGCCCTCAACAAG | CACTCACCTCCATTGGCTGT |
| CrbHLH63 | TGTCCATTGGATGCTCACCC | CAGCACCATCTTGAGTCCGT |
| CrbHLH71 | AGCCACGTACTGCTGAACAA | TCTGCGCTATTGCCAGAACA |
| CrActin | GTCTGTTCCCTCTCCTCCCT | GAAGGGATCGTTGACAGCGA |
Table 1 Primers used for RT-qPCR
| 基因Gene | 上游引物Forward primer(5'-3') | 下游引物Reverse primer(5'-3') |
|---|---|---|
| CrbHLH8 | GCAGTTGTGCCCTCAACAAG | CACTCACCTCCATTGGCTGT |
| CrbHLH13 | CGGCTGAATGCTCGTTGATG | CTGAAATTGGCGGTGTTGGG |
| CrbHLH59 | ACCCCATTGAATTCCCCACC | TACAAATCCAGGATCCGCGG |
| CrbHLH61 | GCAGTTGTGCCCTCAACAAG | CACTCACCTCCATTGGCTGT |
| CrbHLH63 | TGTCCATTGGATGCTCACCC | CAGCACCATCTTGAGTCCGT |
| CrbHLH71 | AGCCACGTACTGCTGAACAA | TCTGCGCTATTGCCAGAACA |
| CrActin | GTCTGTTCCCTCTCCTCCCT | GAAGGGATCGTTGACAGCGA |
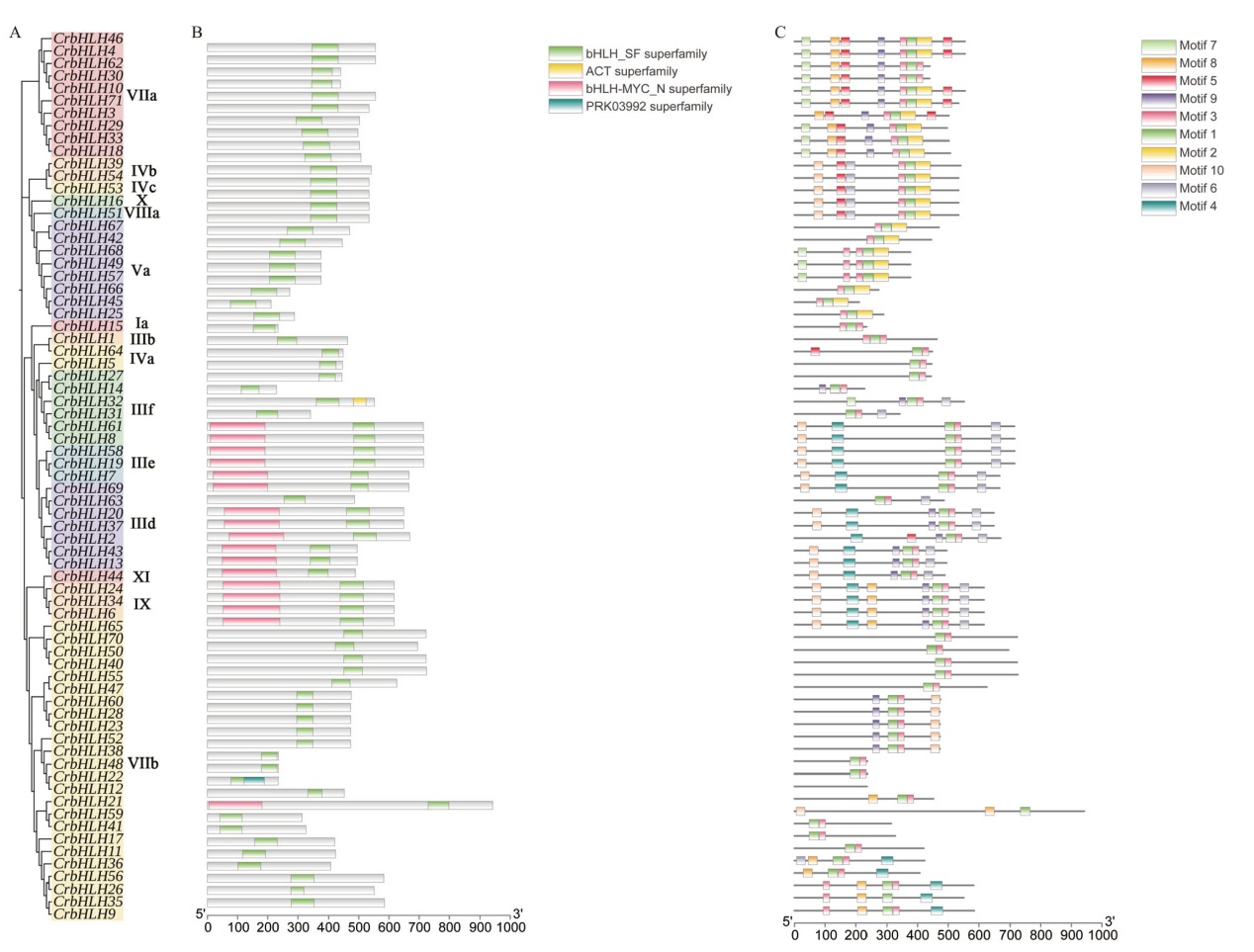
Fig. 3 Analysis of conserved motif and conserved domain of the bHLH gene family of C. reticulate A: Cluster analysis of bHLH gene family members of C. reticulata. B: Conserved motif analysis of bHLH gene members of C. reticulata. C: Conserved domain of the bHLH gene family members of C. reticulata
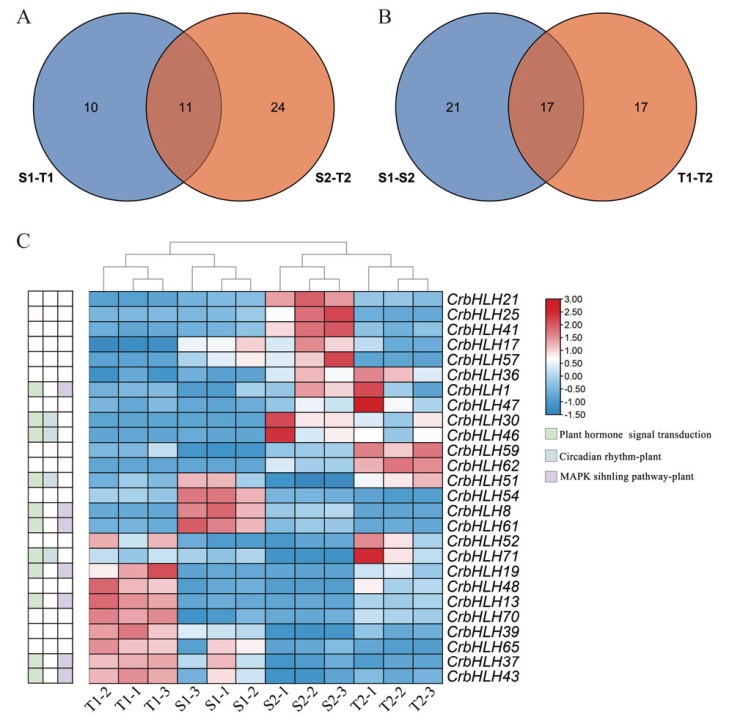
Fig. 4 Differential bHLH analysis of C. reticulate A: Veen map of C. reticulata varieties. B: Veen map of C. reticulata at different developmental stages. C: Differential bHLH gene combination heat map. S: C. reticulata ‘Shizitou’. T: C. reticulata ‘Tongzimian’. 1: Bud stage. 2: Full flowering stage. -1, -2, and -3 indicate three repetitions. The same below
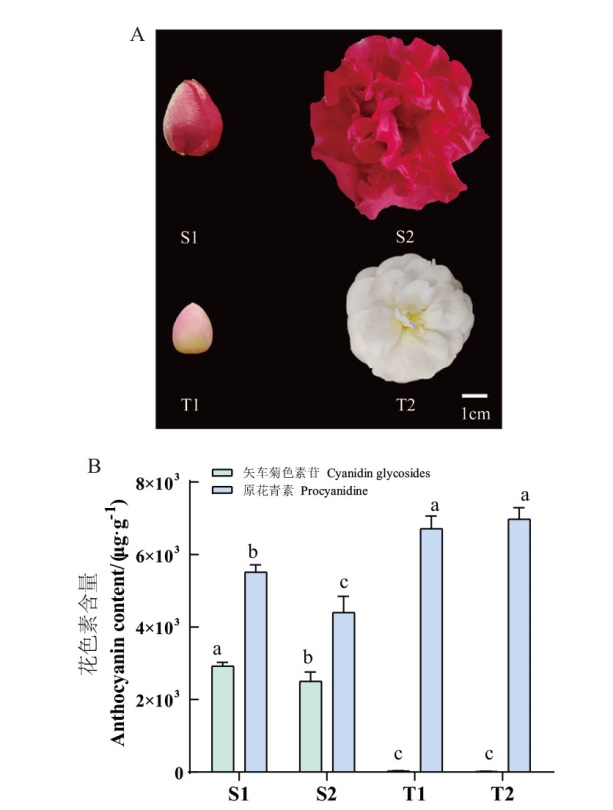
Fig. 5 Floral phenotype(A), and key pigment content(B)in C. reticulata ‘Shizitou’ and C. reticulata ‘Tongzimian’ Different lower letters indicate significant differences at P<0.05 level. The same below
| [1] | 普梅英, 牛锦华, 付芳丽, 等. 云南山茶枝枯病病原菌分离鉴定[J]. 植物病理学报, 2023, 53(6): 1222-1225. |
| Pu MY, Niu JH, Fu FL, et al. Identification of the pathogen causing dieback disease on Camellia reticulata Lindl. in Yunnan[J]. Acta Phytopathol Sin, 2023, 53(6): 1222-1225. | |
| [2] |
吴贵进, 陈龙清, 韦秋雨, 等. 滇山茶登录品种统计及表型性状分析[J]. 园艺学报, 2023, 50(10): 2157-2170.
doi: 10.16420/j.issn.0513-353x.2022-1192 |
| Wu GJ, Chen LQ, Wei QY, et al. Statistics and phenotypic traits analysis of Camellia reticulata registered cultivars[J]. Acta Hortic Sin, 2023, 50(10): 2157-2170. | |
| [3] | Qu Y, Ou Z, Yong QQ, et al. Coloration differences in three Camellia reticulata Lindl. cultivars: ‘Tongzimian’, ‘Shizitou’ and ‘Damanao’[J]. BMC Plant Biol, 2024, 24(1): 18. |
| [4] | Geng F, Nie RM, Yang N, et al. Integrated transcriptome and metabolome profiling of Camellia reticulata reveal mechanisms of flower color differentiation[J]. Front Genet, 2022, 13: 1059717. |
| [5] |
孙晓波, 郭臻昊, 李畅, 等. 杜鹃花品种大鸳鸯锦花瓣条纹形成的色素基础和差异表达基因的挖掘[J]. 华北农学报, 2023, 38(6): 26-35.
doi: 10.7668/hbnxb.20194067 |
| Sun XB, Guo ZH, Li C, et al. The pigment foundation of the formation of petal stripes in the Rhododendron variety dayuanyangjin and the exploration of differential expression genes[J]. Acta Agric Boreali Sin, 2023, 38(6): 26-35. | |
| [6] | 黄虎, 马先进, 李思佳, 等. 山茶CjMYB1基因的克隆及表达分析[J]. 西北植物学报, 2022, 42(3): 381-389. |
| Huang H, Ma XJ, Li SJ, et al. Molecular cloning and expression analysis of CjMYB1 in Camellia japonica[J]. Acta Bot Boreali Occidentalia Sin, 2022, 42(3): 381-389. | |
| [7] | Li JB, Hashimoto F, Shimizu K, et al. Anthocyanins from red flowers of Camellia reticulata LINDL[J]. Biosci Biotechnol Biochem, 2007, 71(11): 2833-2836. |
| [8] |
安昌, 陆琳, 沈梦千, 等. 植物bHLH基因家族研究进展及在药用植物中的应用前景[J]. 生物技术通报, 2023, 39(10): 1-16.
doi: 10.13560/j.cnki.biotech.bull.1985.2023-0243 |
| An C, Lu L, Shen MQ, et al. Research progress of bHLH gene family in plants and its application prospects in medical plants[J]. Biotechnol Bull, 2023, 39(10): 1-16. | |
| [9] | 杨鹏程, 周波, 李玉花. 植物花青素合成相关的bHLH转录因子[J]. 植物生理学报, 2012, 48(8): 747-758. |
| Yang PC, Zhou B, Li YH. The bHLH transcription factors involved in anthocyanin biosynthesis in plants[J]. Plant Physiol J, 2012, 48(8): 747-758. | |
| [10] | Zhang KK, Lin CY, Chen BY, et al. A light responsive transcription factor CsbHLH89 positively regulates anthocyanidin synthesis in tea(Camellia sinensis)[J]. Sci Hortic, 2024, 327: 112784. |
| [11] |
李博, 刘合霞, 陈宇玲, 等. 金花茶CnbHLH79转录因子的克隆、亚细胞定位及表达分析[J]. 生物技术通报, 2023, 39(8): 241-250.
doi: 10.13560/j.cnki.biotech.bull.1985.2022-1562 |
| Li B, Liu HX, Chen YL, et al. Cloning, subcellular localization and expression analysis of CnbHLH79 transcription factor from Camellia nitidissima[J]. Biotechnol Bull, 2023, 39(8): 241-250. | |
| [12] |
Schaart JG, Dubos C, Romero De La Fuente I, et al. Identification and characterization of MYB-bHLH-WD40 regulatory complexes controlling proanthocyanidin biosynthesis in strawberry(Fragaria × ananassa)fruits[J]. New Phytol, 2013, 197(2): 454-467.
doi: 10.1111/nph.12017 pmid: 23157553 |
| [13] |
Koramutla MK, Ram C, Bhatt D, et al. Genome-wide identification and expression analysis of sucrose synthase genes in allotetraploid Brassica juncea[J]. Gene, 2019, 707: 126-135.
doi: S0378-1119(19)30417-2 pmid: 31026572 |
| [14] | An JP, Zhang XW, Liu YJ, et al. ABI5 regulates ABA-induced anthocyanin biosynthesis by modulating the MYB1-bHLH3 complex in apple[J]. J Exp Bot, 2021, 72(4): 1460-1472. |
| [15] | Shen ZG, Li WY, Li YL, et al. The red flower wintersweet genome provides insights into the evolution of magnoliids and the molecular mechanism for tepal color development[J]. Plant J, 2021, 108(6): 1662-1678. |
| [16] |
Heim MA, Jakoby M, Werber M, et al. The basic helix-loop-helix transcription factor family in plants: a genome-wide study of protein structure and functional diversity[J]. Mol Biol Evol, 2003, 20(5): 735-747.
doi: 10.1093/molbev/msg088 pmid: 12679534 |
| [17] |
葛诗蓓, 张学宁, 韩文炎, 等. 植物类黄酮的生物合成及其抗逆作用机制研究进展[J]. 园艺学报, 2023, 50(1): 209-224.
doi: 10.16420/j.issn.0513-353x.2021-1186 |
|
Ge SB, Zhang XN, Han WY, et al. Research progress on plant flavonoids biosynthesis and their anti-stress mechanism[J]. Acta Hortic Sin, 2023, 50(1): 209-224.
doi: 10.16420/j.issn.0513-353x.2021-1186 |
|
| [18] |
贺威智, 雷伟奇, 郭祥鑫, 等. 百子莲MYB家族鉴定及蓝色形成关键基因功能分析[J]. 园艺学报, 2023, 50(6): 1255-1268.
doi: 10.16420/j.issn.0513-353x.2022-0226 |
| He WZ, Lei WQ, Guo XX, et al. Identification of the MYB gene family and functional analysis of key genes related to blue flower coloration in Agapanthus praecox[J]. Acta Hortic Sin, 2023, 50(6): 1255-1268. | |
| [19] | 龙芬芳. 马缨杜鹃花色苷合成相关MYB基因的筛选与功能初步验证[D]. 贵阳: 贵州大学, 2023: 1-5. |
| Long FF. Screening and preliminary functional verification of MYB gene related to anthocyanin synthesis in Rhododendron camelliottii[D]. Guiyang: Guizhou University, 2023: 1-5. | |
| [20] | 邹红竹, 韩璐璐, 周琳, 等. 滇牡丹MYB家族成员鉴定及PdMYB2的功能验证[J]. 林业科学研究, 2022, 35(5): 1-13. |
| Zou HZ, Han LL, Zhou L, et al. Identification of MYB family members and functional verification of PdMYB2 in Paeonia delavayi[J]. For Res, 2022, 35(5): 1-13. | |
| [21] |
范文广, 李保豫, 田辉, 等. 兰州百合鳞茎花青素合成相关MYB与bHLH转录因子的筛选分析[J]. 食品与发酵工业, 2024, 50(2): 57-66.
doi: 10.13995/j.cnki.11-1802/ts.033270 |
| Fan WG, Li BY, Tian H, et al. Screening and analysis of MYB and bHLH transcription factors associated with anthocyanin synthesis in Lilium davidii var. unicolor bulbs[J]. Food Ferment Ind, 2024, 50(2): 57-66. | |
| [22] | 王洪飞, 欧静, 王孝敬, 等. 马缨杜鹃bHLH转录因子家族的鉴定与表达分析[J/OL]. 广西植物, 2023. https://kns.cnki.net/kcms/detail/45.1134.Q.20230720.2036.002.html. |
| Wang HF, Ou J, Wang XJ, et al. Identification and expression analysis of bHLH transcription factors family in Rhododendron delavayi[J/OL]. Guihaia, 2023. https://kns.cnki.net/kcms/detail/45.1134.Q.20230720.2036.002.html. | |
| [23] | 陈丽飞, 刘云怡慧, 李嘉峻, 等. 干旱胁迫下大苞萱草bHLH转录因子家族鉴定与分析[J]. 西南农业学报, 2023, 36(5): 1027-1038. |
| Chen LF, Liu Y, Li JJ, et al. Identification and analysis of bHLH transcription factor gene family in Hemerocallis middendorffii under drought stress[J]. Southwest China J Agric Sci, 2023, 36(5): 1027-1038. | |
| [24] | Li XJ, Cao LJ, Jiao BB, et al. The bHLH transcription factor AcB2 regulates anthocyanin biosynthesis in onion(Allium cepa L.)[J]. Hortic Res, 2022, 9: uhac128. |
| [25] | 陈佳红, 蒋玲莉, 钱婕妤, 等. 百日草bHLH家族IIIf亚族基因筛选及表达分析[J]. 农业生物技术学报, 2023, 31(1): 61-72. |
| Chen JH, Jiang LL, Qian JY, et al. Screening and expression analysis of IIIf subgroup gene of bHLH family in Zinnia elegans[J]. J Agric Biotechnol, 2023, 31(1): 61-72. | |
| [26] | Song QL, Gong WF, Yu XR, et al. Transcriptome and anatomical comparisons reveal the effects of methyl jasmonate on the seed development of Camellia oleifera[J]. J Agric Food Chem, 2023, 71(17): 6747-6762. |
| [27] |
Dombrecht B, Xue GP, Sprague SJ, et al. MYC2 differentially modulates diverse jasmonate-dependent functions in Arabidopsis[J]. Plant Cell, 2007, 19(7): 2225-2245.
doi: 10.1105/tpc.106.048017 pmid: 17616737 |
| [28] |
Lorenzo O, Chico JM, Sánchez-Serrano JJ, et al. JASMONATE-INSENSITIVE1 encodes a MYC transcription factor essential to discriminate between different jasmonate-regulated defense responses in Arabidopsis[J]. Plant Cell, 2004, 16(7): 1938-1950.
doi: 10.1105/tpc.022319 pmid: 15208388 |
| [29] | Schweizer F, Fernández-Calvo P, Zander M, et al. Arabidopsis basic helix-loop-helix transcription factors MYC2, MYC3, and MYC 4 regulate glucosinolate biosynthesis, insect performance, and feeding behavior[J]. Plant Cell, 2013, 25(8): 3117-3132. |
| [30] | 周麟, 雍清青, 姚响, 等. 滇山茶狮子头及其芽变品种大玛瑙间花色差异分析[J]. 植物遗传资源学报, 2024, 25(4): 960-971. |
| Zhou L, Yong QQ, Yao X, et al. Analysis of flower color difference between Camellia reticulata ‘shizitou’ and its bud mutant ‘damanao’[J]. J Plant Genet Resour, 2024, 25(4): 960-971. | |
| [31] | Qi Y, Zhou L, Han LL, et al. PsbHLH1, a novel transcription factor involved in regulating anthocyanin biosynthesis in tree peony(Paeonia suffruticosa)[J]. Plant Physiol Biochem, 2020, 154: 396-408. |
| [32] |
Tao RY, Yu WJ, Gao YH, et al. Light-induced basic/helix-loop-helix64 enhances anthocyanin biosynthesis and undergoes CONSTITUTIVELY PHOTOMORPHOGENIC1-mediated degradation in pear[J]. Plant Physiol, 2020, 184(4): 1684-1701.
doi: 10.1104/pp.20.01188 pmid: 33093233 |
| [33] |
吕秋谕, 孙培媛, 冉彬, 等. 苦荞转录因子基因FtbHLH3的克隆、亚细胞定位及表达分析[J]. 生物技术通报, 2023, 39(8): 194-203.
doi: 10.13560/j.cnki.biotech.bull.1985.2023-0034 |
| Lyu QY, Sun PY, Ran B, et al. Cloning, subcellular localization and expression analysis of the transcription factor gene FtbHLH3 in Fagopyrum tataricum[J]. Biotechnol Bull, 2023, 39(8): 194-203. |
| [1] | YANG Wei, ZHAO Li-fen, TANG Bing, ZHOU Lin-bi, YANG Juan, MO Chuan-yuan, ZHANG Bao-hui, LI Fei, RUAN Song-lin, DENG Ying. Genome-wide Identification and Expression Analysis of the SRO Gene Family in Brassica juncea L. [J]. Biotechnology Bulletin, 2024, 40(8): 129-141. |
| [2] | WU Shuai, XIN Yan-ni, MAI Chun-hai, MU Xiao-ya, WANG Min, YUE Ai-qin, ZHAO Jin-zhong, WU Shen-jie, DU Wei-jun, WANG Li-xiang. Genome-wide Identification and Stress Response Analysis of Soybean GS Gene Family [J]. Biotechnology Bulletin, 2024, 40(8): 63-73. |
| [3] | ZHANG Ming-ya, PANG Sheng-qun, LIU Yu-dong, SU Yong-feng, NIU Bo-wen, HAN Qiong-qiong. Identification and Expression Analysis of FAD Gene Family in Solanum lycopersicum [J]. Biotechnology Bulletin, 2024, 40(7): 150-162. |
| [4] | ZANG Wen-rui, MA Ming, CHE Gen, HASI Agula. Genome-wide Identification and Expression Pattern Analysis of BZR Transcription Factor Gene Family of Melon [J]. Biotechnology Bulletin, 2024, 40(7): 163-171. |
| [5] | WANG Jian, YANG Sha, SUN Qing-wen, CHEN Hong-yu, YANG Tao, HUANG Yuan. Genome-wide Identification and Expression Analysis of bHLH Transcription Factor Family in Dendrobium nobile [J]. Biotechnology Bulletin, 2024, 40(6): 203-218. |
| [6] | LI Meng-ran, YE Wei, LI Sai-ni, ZHANG Wei-yang, LI Jian-jun, ZHANG Wei-min. Expression of Lithocarols Biosynthesis Gene litI and Functional Analysis of Its Promoter [J]. Biotechnology Bulletin, 2024, 40(6): 310-318. |
| [7] | HU Yong-bo, LEI Yu-tian, YANG Yong-sen, CHEN Xin, LIN Huang-fang, LIN Bi-ying, LIU Shuang, BI Ge, SHEN Bao-ying. Genome-wide Identification and Expression Pattern Analysis of the Bcl-2-related Anti-apoptotic Family in Cucumis sativus L. and Cucurbita moschata Duch. [J]. Biotechnology Bulletin, 2024, 40(6): 219-237. |
| [8] | CHANG Xue-rui, WANG Tian-tian, WANG Jing. Identification and Analysis of E2 Gene Family in Pepper(Capsicum annuum L.) [J]. Biotechnology Bulletin, 2024, 40(6): 238-250. |
| [9] | LIU Rong, TIAN Min-yu, LI Guang-ze, TAN Cheng-fang, RUAN Ying, LIU Chun-lin. Identification and Induced-expression Analysis of REVEILLE Family in Brassica napus L. [J]. Biotechnology Bulletin, 2024, 40(6): 161-171. |
| [10] | HOU Ya-qiong, LANG Hong-shan, WEN Meng-meng, GU Yi-yun, ZHU Run-jie, TANG Xiao-li. Identification and Expression Analysis of AcHSP20 Gene Family in Kiwifruit [J]. Biotechnology Bulletin, 2024, 40(5): 167-178. |
| [11] | LI Jia-xin, LI Hong-yan, LIU Li-e, ZHANG Tian, ZHOU Wu. Identification and Expression Analysis of the NRAMP Family in Seabuckthorn Under Lead Stress [J]. Biotechnology Bulletin, 2024, 40(5): 191-202. |
| [12] | XIAO Ya-ru, JIA Ting-ting, LUO Dan, WU Zhe, LI Li-xia. Cloning and Expression Analysis of CsERF025L Transcription Factor in Cucumber [J]. Biotechnology Bulletin, 2024, 40(4): 159-166. |
| [13] | ZHONG Yun, LIN Chun, LIU Zheng-jie, DONG Chen-wen-hua, MAO Zi-chao, LI Xing-yu. Cloning and Prokaryotic Expression Analysis of Asparagus Saponin Synthesis Related Glycosyltransferase Genes [J]. Biotechnology Bulletin, 2024, 40(4): 255-263. |
| [14] | ZHANG Qing-lan, ZHANG Ya-ran, JU Zhi-hua, WANG Xiu-ge, XIAO Yao, WANG Jin-peng, WEI Xiao-chao, GAO Ya-ping, BAI Fu-heng, WANG Hong-cheng. Identification and Transcriptional Regulation Analysis of Core Promoter in Bovine TARDBP Gene [J]. Biotechnology Bulletin, 2024, 40(4): 306-318. |
| [15] | CHEN Xiao-song, LIU Chao-jie, ZHENG Jia, QIAO Zong-wei, LUO Hui-bo, ZOU Wei. Analyzing the Growth and Caproic Acid Metabolism Mechanism of Rummeliibacillus suwonensis 3B-1 by Tandem Mass Tag-based Quantitative Proteomics [J]. Biotechnology Bulletin, 2024, 40(3): 135-145. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||