








Biotechnology Bulletin ›› 2025, Vol. 41 ›› Issue (3): 14-24.doi: 10.13560/j.cnki.biotech.bull.1985.2024-1038
Previous Articles Next Articles
LU Feng1( ), HUANG Yu-hong2, LIN Yan-na3, MA Fu-qiang1(
), HUANG Yu-hong2, LIN Yan-na3, MA Fu-qiang1( )
)
Received:2024-10-24
Online:2025-03-26
Published:2025-03-20
Contact:
MA Fu-qiang
E-mail:luf@sibet.ac.cn;mafuqiang318@sibet.ac.cn
LU Feng, HUANG Yu-hong, LIN Yan-na, MA Fu-qiang. Advances on Molecular Modifications of Formate Dehydrogenase for CO₂ Reduction[J]. Biotechnology Bulletin, 2025, 41(3): 14-24.

Fig. 1 Flowchart of multi-enzyme cascade catalysis for CO2 to methanol synthesisFDH: Formate dehydrogenase; FaldDH: formaldehyde dehydrogenase; ADH: alcohol dehydrogenase; NADH: reduced nicotinamide adenine dinucleotide; NAD+: oxidized nicotinamide adenine dinucleotide; W/Mo: tungsten/molybdenum
| 类型 Types | 物种 Species | FDH | 分子量 Mw /kD | kcat /(s-1) | KM /(mmol·L-1) | kcat/KM / (mmol·L-1·s-1) | 参考文献 Reference |
|---|---|---|---|---|---|---|---|
| NAD+依赖型FDH | Candida boidinii | CbFDH | 41.0 | 0.015 0 | 31.3 | 0.000 400 | [ |
| Thiobacillus sp. KNK65MA | TsFDH | 45.0 | 0.318 | 9.23 | 0.034 0 | [ | |
| Thermochaetoides thermophila DSM 1495 | CtFDH | 45.0 | 0.023 0 | 0.320 | 0.069 0 | [ | |
| Candida methylica | CmFDH | 42.0 | 0.008 00 | 0.780 | 0.010 0 | [ | |
| Thermothelomyces thermophilus ATCC 42464 | MtFDH | 42.0 | 0.100 | 0.400 | 0.250 | [ | |
| Paracoccus sp. MKU1 | PsFDH | 44.0 | 0.073 0 | 0.928 | 0.079 0 | [ | |
| 金属依赖型FDH | Desulfovibrio desulfuricans | DdFDH | 135 | 46.6 | 0.015 7 | 2.97×103 | [ |
| Desulfovibrio vulgaris Hildenborough | DvFdhAB | 97.4 | 315 | 0.420 | 750 | [ | |
| Cupriavidus necator | FdsABG | 178 | 11.0 | 2.70 | 4.07 | [ | |
| Acetobacterium woodii | FdhF1/2 | 169 | 372 | 3.80 | 97.9 | [ | |
| Escherichia coli | EcFDH-H | 79.0 | 1.00 | 8.30 | 0.120 | [ | |
| Clostridium Ijungdahlii | ClFDH | 80.0 | 0.012 0 | 7.27 | 0.001 65 | [ | |
| Clostridium ljungdahlii | ClFDH | 75.0 | 5.66 | 66.2 | 0.085 5 | [ | |
| Clostridium autoethanogenum | CaFDH | 75.0 | 4.00 | 23.2 | 0.170 | [ | |
| Clostridium coskatii | CcFDH | 75.0 | 5.62 | 59.7 | 0.094 0 | [ | |
| Clostridium ragsdalei | CrFDH | 75.0 | 3.28 | 31.2 | 0.110 | [ | |
| Clostridium carboxidivorans P7T | FDHH_CloCa | 80.7 | 0.080 0 | 0.050 0 | 1.60 | [ | |
| Pseudomonas oxalaticus | Formate Dehydrogenase | 315 | 3.00 | 40.0 | 0.075 0 | [ | |
| Desulfosporosinus acididurans | DaFDH | 93.0 | 4.09 | 34.9 | 0.117 | [ | |
| Paraclostridium bifermentans | PbFDH | 93.0 | 4.45 | 30.6 | 0.145 | [ | |
| Clostridium scatologenes | CsFDH | 79.0 | 1.55 | 48.9 | 0.031 7 | [ |
Table 1 Types and kinetic parameters of FDH for CO2 reduction
| 类型 Types | 物种 Species | FDH | 分子量 Mw /kD | kcat /(s-1) | KM /(mmol·L-1) | kcat/KM / (mmol·L-1·s-1) | 参考文献 Reference |
|---|---|---|---|---|---|---|---|
| NAD+依赖型FDH | Candida boidinii | CbFDH | 41.0 | 0.015 0 | 31.3 | 0.000 400 | [ |
| Thiobacillus sp. KNK65MA | TsFDH | 45.0 | 0.318 | 9.23 | 0.034 0 | [ | |
| Thermochaetoides thermophila DSM 1495 | CtFDH | 45.0 | 0.023 0 | 0.320 | 0.069 0 | [ | |
| Candida methylica | CmFDH | 42.0 | 0.008 00 | 0.780 | 0.010 0 | [ | |
| Thermothelomyces thermophilus ATCC 42464 | MtFDH | 42.0 | 0.100 | 0.400 | 0.250 | [ | |
| Paracoccus sp. MKU1 | PsFDH | 44.0 | 0.073 0 | 0.928 | 0.079 0 | [ | |
| 金属依赖型FDH | Desulfovibrio desulfuricans | DdFDH | 135 | 46.6 | 0.015 7 | 2.97×103 | [ |
| Desulfovibrio vulgaris Hildenborough | DvFdhAB | 97.4 | 315 | 0.420 | 750 | [ | |
| Cupriavidus necator | FdsABG | 178 | 11.0 | 2.70 | 4.07 | [ | |
| Acetobacterium woodii | FdhF1/2 | 169 | 372 | 3.80 | 97.9 | [ | |
| Escherichia coli | EcFDH-H | 79.0 | 1.00 | 8.30 | 0.120 | [ | |
| Clostridium Ijungdahlii | ClFDH | 80.0 | 0.012 0 | 7.27 | 0.001 65 | [ | |
| Clostridium ljungdahlii | ClFDH | 75.0 | 5.66 | 66.2 | 0.085 5 | [ | |
| Clostridium autoethanogenum | CaFDH | 75.0 | 4.00 | 23.2 | 0.170 | [ | |
| Clostridium coskatii | CcFDH | 75.0 | 5.62 | 59.7 | 0.094 0 | [ | |
| Clostridium ragsdalei | CrFDH | 75.0 | 3.28 | 31.2 | 0.110 | [ | |
| Clostridium carboxidivorans P7T | FDHH_CloCa | 80.7 | 0.080 0 | 0.050 0 | 1.60 | [ | |
| Pseudomonas oxalaticus | Formate Dehydrogenase | 315 | 3.00 | 40.0 | 0.075 0 | [ | |
| Desulfosporosinus acididurans | DaFDH | 93.0 | 4.09 | 34.9 | 0.117 | [ | |
| Paraclostridium bifermentans | PbFDH | 93.0 | 4.45 | 30.6 | 0.145 | [ | |
| Clostridium scatologenes | CsFDH | 79.0 | 1.55 | 48.9 | 0.031 7 | [ |

Fig. 2 Structure and catalytic mechanism of CbFDHA: Three-dimensional structure of CbFDH (PDB: 6D4C). The two dimers are colored in red and green, while the NAD+ cofactor is colored in orange. B: Active center of CbFDH and amino acid residues closed to NAD+. C: Mechanism of action of NAD+-dependent FDH for formate oxidation (green arrows) or CO2 reduction (inverse sense, pink arrows)
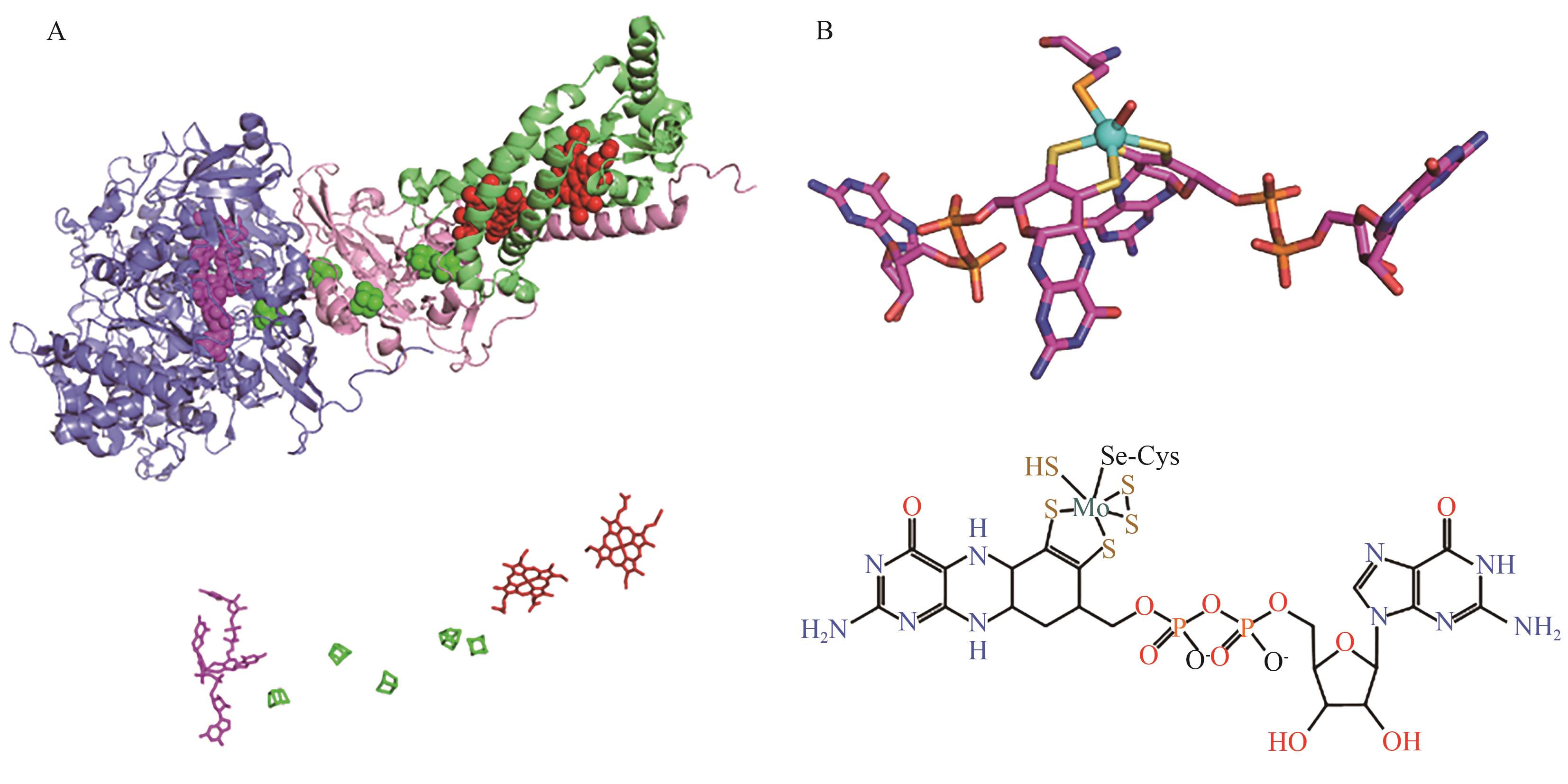
Fig. 3 Three-dimensional structure and cofactor diagram of E. coli FDH NA: Three-dimensional structure of Escherichia coli FDH N (PDB: 1KQF). The three polypeptide chains are indicated by blue (chain A), pink (chain B) and green (chain C). The molybdopterin (in chain A), the five [4Fe-4S] clusters (in chain A and chain B) and the two heme (chain C) are displayed in pink, green and red, respectively. B: Graphic of the molybdenum cofactor and its chemical formula structure in E. coli FDH N
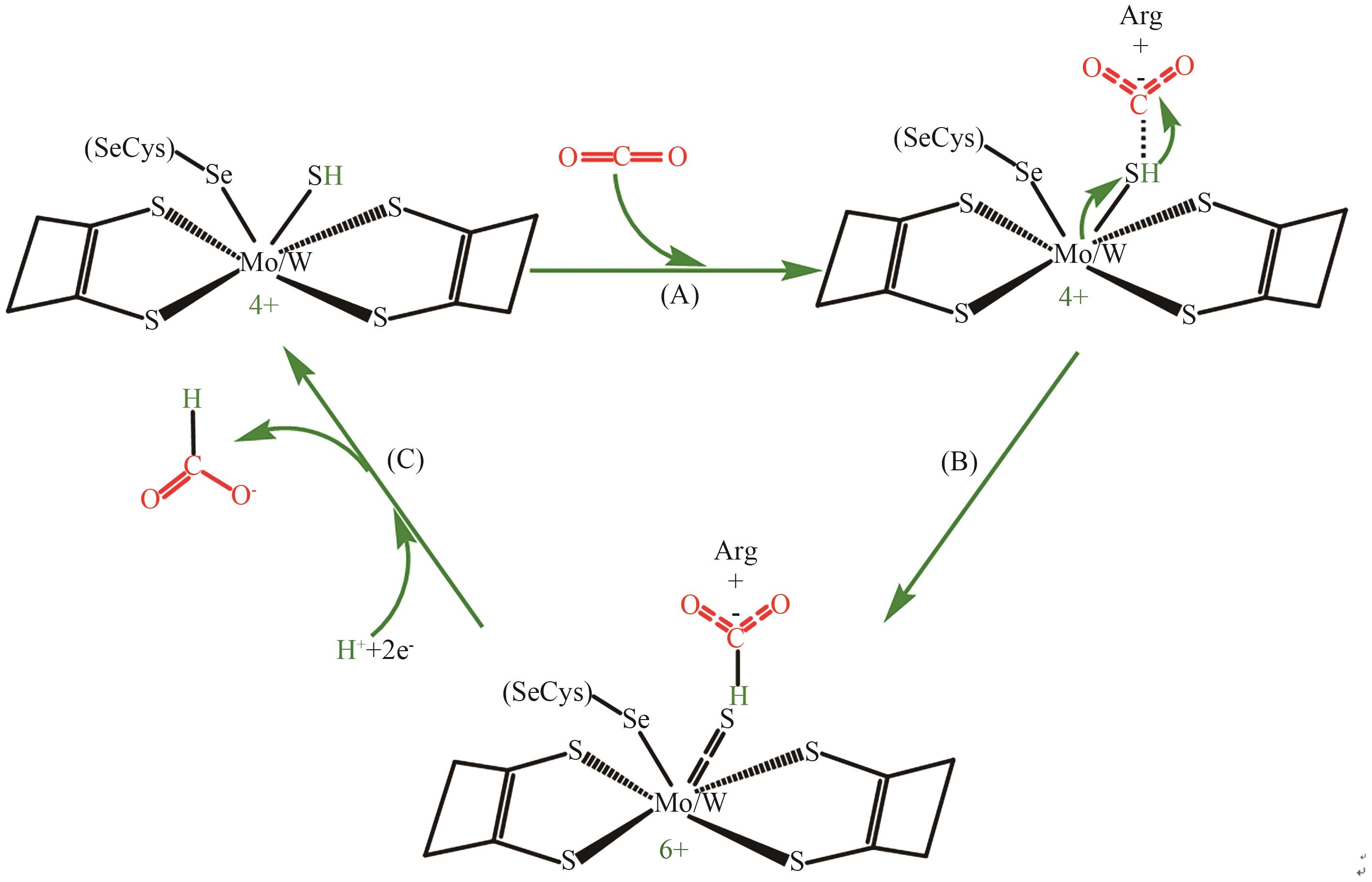
Fig. 4 Feasible mechanism of CO2 reduction to formic acid with metal-dependent FDHA: CO2 is electrostatically captured by positively charged arginine residues of FDH. B: Hydrogen mercaptan bound to metal Mo/W attacks CO2 captured by arginine residues, and the valence state is oxidized from four to six. C: Under the action of accepting proton electrons and releasing formic acid, the valence state of metal Mo/W is reduced from six to four, and the initial state is restored
| 35 | Alpdagtas S, Binay B. NADP+-dependent formate dehydrogenase: a review [J]. Biocatal Biotransform, 2021, 39(4): 260-268. |
| 36 | Alpdağtaş S, Turunen O, Valjakka J, et al. The challenges of using NAD+-dependent formate dehydrogenases for CO2 conversion [J]. Crit Rev Biotechnol, 2022, 42(6): 953-972. |
| 37 | Castillo R, Oliva M, Martí S, et al. A theoretical study of the catalytic mechanism of formate dehydrogenase [J]. J Phys Chem B, 2008, 112(32): 10012-10022. |
| 38 | Çakar MM, Ruupunen J, Mangas-Sanchez J, et al. Engineered formate dehydrogenase from Chaetomium thermophilum, a promising enzymatic solution for biotechnical CO2 fixation [J]. Biotechnol Lett, 2020, 42(11): 2251-2262. |
| 39 | Meneghello M, Oliveira AR, Jacq-Bailly A, et al. Formate dehydrogenases reduce CO2 rather than HCO3 -: an electrochemical demonstration [J]. Angew Chem Int Ed, 2021, 60(18): 9964-9967. |
| 40 | Pagano P, Guo Q, Ranasinghe C, et al. Oscillatory active-site motions correlate with kinetic isotope effects in formate dehydrogenase [J]. ACS Catal, 2019, 9(12): 11199-11206. |
| 41 | Cordas CM, Moura JJG. Molybdenum and tungsten enzymes redox properties-A brief overview [J]. Coord Chem Rev, 2019, 394: 53-64. |
| 42 | Kirk ML, Hille R. Spectroscopic studies of mononuclear molybdenum enzyme centers [J]. Molecules, 2022, 27(15): 4802. |
| 43 | Jormakka M, Törnroth S, Byrne B, et al. Molecular basis of proton motive force generation: structure of formate dehydrogenase-N [J]. Science, 2002, 295(5561): 1863-1868. |
| 44 | Maia LB, Moura I, Moura JJG. Molybdenum and tungsten-containing formate dehydrogenases: aiming to inspire a catalyst for carbon dioxide utilization [J]. Inorg Chim Acta, 2017, 455: 350-363. |
| 45 | Guo Q, Gakhar L, Wickersham K, et al. Structural and kinetic studies of formate dehydrogenase from Candida boidinii [J]. Biochemistry, 2016, 55(19): 2760-2771. |
| 46 | Sato R, Amao Y. Studies on the catalytic mechanism of formate dehydrogenase from Candida boidinii using isotope-labelled substrate and co-enzyme [J]. Catal Today, 2023, 411: 113796. |
| 47 | Jiang W, Lin P, Yang RN, et al. Identification of catalysis, substrate, and coenzyme binding sites and improvement catalytic efficiency of formate dehydrogenase from Candida boidinii [J]. Appl Microbiol Biotechnol, 2016, 100(19): 8425-8437. |
| 48 | Tishkov VI, Popov VO. Protein engineering of formate dehydrogenase [J]. Biomol Eng, 2006, 23(2/3): 89-110. |
| 49 | Pala U, Yelmazer B, Çorbacıoğlu M, et al. Functional effects of active site mutations in NAD+-dependent formate dehydrogenases on transformation of hydrogen carbonate to formate [J]. Protein Eng Des Sel, 2018, 31(9): 327-335. |
| 50 | Tülek A, Günay E, Servili B, et al. Sustainable production of formic acid from CO2 by a novel immobilized mutant formate dehydrogenase [J]. Sep Purif Technol, 2023, 309: 123090. |
| 51 | Kurt S, Ordu E. Effect of Met/Leu substitutions on the stability of NAD+-dependent formate dehydrogenases from Gossypium hirsutum [J]. Appl Microbiol Biotechnol, 2021, 105(7): 2787-2798. |
| 52 | Shi HL, Fu MR, Zhang TT, et al. Rational design of formate dehydrogenase for enhanced thermal stability and catalytic activity in bioelectrocatalysis [J]. J Agric Food Chem, 2024, 72(42): 23333-23344. |
| 53 | Schirwitz K, Schmidt A, Lamzin VS. High-resolution structures of formate dehydrogenase from Candida boidinii [J]. Protein Sci, 2007, 16(6): 1146-1156. |
| 54 | Alekseeva AA, Fedorchuk VV, Zarubina SA, et al. The role of Ala198 in the stability and coenzyme specificity of bacterial formate dehydrogenases [J]. Acta Naturae, 2015, 7(1): 60-69. |
| 55 | Calzadiaz-Ramirez L, Calvó-Tusell C, Stoffel GMM, et al. In vivo selection for formate dehydrogenases with high efficiency and specificity toward NADP+ [J]. ACS Catal, 2020, 10(14): 7512-7525. |
| 56 | Fogal S, Beneventi E, Cendron L, et al. Structural basis for double cofactor specificity in a new formate dehydrogenase from the Acidobacterium Granulicella mallensis MP5ACTX8 [J]. Appl Microbiol Biotechnol, 2015, 99(22): 9541-9554. |
| 57 | Robescu DMS, Rubini R, Beneventi DE, et al. From the amelioration of a NADP+-dependent formate dehydrogenase to the discovery of a new enzyme: round trip from theory to practice [J]. ChemCatChem, 2020, 12(9): 2478-2487. |
| 58 | Guo XJ, Wang XY, Liu YX, et al. Structure-guided design of formate dehydrogenase for regeneration of a non-natural redox cofactor [J]. Chemistry, 2020, 26(70): 16611-16615. |
| 1 | Jia ZC, Dang JN, Wen GB, et al. Constructing nanocaged enzymes for synergistic catalysis of CO2 reduction [J]. Adv Sci, 2023, 10(20): e2300752. |
| 2 | Chen H, Huang Y, Sha C, et al. Enzymatic carbon dioxide to formate: mechanisms, challenges and opportunities [J]. Renew Sustain Energy Rev, 2023, 178: 113271. |
| 3 | IEA. CO2 Emissions in 2023[EB/OL]. (2024-03-01) [2024-10-24]. . |
| 4 | Yuan SW, Fu MR, Xian YM, et al. Efficient heterologous expression of formate dehydrogenase and preliminary determination of the potential for conversion of carbon dioxide to formate [J]. Mol Catal, 2023, 548: 113455. |
| 5 | Li YX, Yan LH, Liu GH, et al. Enhanced electroenzymatic CO2 reduction by a multifunctional ZIF-8 layer on silica nanoflower with immobilized enzyme [J]. Chem Eng J, 2023, 466: 143198. |
| 6 | Cai T, Sun HB, Qiao J, et al. Cell-free chemoenzymatic starch synthesis from carbon dioxide [J]. Science, 2021, 373(6562): 1523-1527. |
| 7 | Ji XL, Guo H, Xue YJ, et al. Microenvironment: an efficient avenue for converting CO2 to high-value compounds [J]. Renew Sustain Energy Rev, 2023, 188: 113809. |
| 8 | Moon M, Park GW, Lee JP, et al. Recent progress in formate dehydrogenase (FDH) as a non-photosynthetic CO2 utilizing enzyme: a short review [J]. J CO2 Util, 2020, 42: 101353. |
| 9 | Villa R, Nieto S, Donaire A, et al. Direct biocatalytic processes for CO2 capture as a green tool to produce value-added chemicals [J]. Molecules, 2023, 28(14): 5520. |
| 10 | Kuwabata S, Tsuda R, Nishida K, et al. Electrochemical conversion of carbon dioxide to methanol with use of enzymes as biocatalysts [J]. Chem Lett, 1993, 22(9): 1631-1634. |
| 11 | Calzadiaz-Ramirez L, Meyer AS. Formate dehydrogenases for CO2 utilization [J]. Curr Opin Biotechnol, 2022, 73: 95-100. |
| 12 | Nielsen CF, Lange L, Meyer AS. Classification and enzyme kinetics of formate dehydrogenases for biomanufacturing via CO2 utilization [J]. Biotechnol Adv, 2019, 37(7): 107408. |
| 13 | Hille R, Hall J, Basu P. The mononuclear molybdenum enzymes [J]. Chem Rev, 2014, 114(7): 3963-4038. |
| 14 | Popov VO, Lamzin VS. NAD+-dependent formate dehydrogenase [J]. Biochem J, 1994, 301 (Pt 3)(Pt 3): 625-643. |
| 15 | Jollie DR, Lipscomb JD. Formate dehydrogenase from Methylosinus trichosporium OB3b. Purification and spectroscopic characterization of the cofactors [J]. J Biol Chem, 1991, 266(32): 21853-21863. |
| 16 | Amao Y. Formate dehydrogenase for CO2 utilization and its application [J]. J CO2 Util, 2018, 26: 623-641. |
| 17 | Oliveira AR, Mota C, Mourato C, et al. Toward the mechanistic understanding of enzymatic CO2 reduction [J]. ACS Catal, 2020, 10(6): 3844-3856. |
| 18 | Shi HL, Fu MR, Yuan SW, et al. Engineered Escherichia coli whole cell-mediated electro-biocatalysis for carbon dioxide to formic acid conversion [J]. ACS Sustainable Chem Eng, 2024, 12(14): 5544-5554. |
| 19 | Min K, Park YS, Park GW, et al. Elevated conversion of CO2 to versatile formate by a newly discovered formate dehydrogenase from Rhodobacter aestuarii [J]. Bioresour Technol, 2020, 305: 123155. |
| 20 | Choe H, Joo JC, Cho DH, et al. Efficient CO2-reducing activity of NAD-dependent formate dehydrogenase from Thiobacillus sp. KNK65MA for formate production from CO2 gas [J]. PLoS One, 2014, 9(7): e103111. |
| 21 | Aslan AS, Valjakka J, Ruupunen J, et al. Chaetomium thermophilum formate dehydrogenase has high activity in the reduction of hydrogen carbonate (HCO3 -) to formate [J]. Protein Eng Des Sel, 2017, 30(1): 47-55. |
| 22 | Altaş N, Aslan AS, Karataş E, et al. Heterologous production of extreme alkaline thermostable NAD+-dependent formate dehydrogenase with wide-range pH activity from Myceliophthora thermophila [J]. Process Biochem, 2017, 61: 110-118. |
| 23 | Xue YJ, Ji XL, Li Z, et al. NADH-dependent formate dehydrogenase mutants for efficient carbon dioxide fixation [J]. Bioresour Technol, 2024, 393: 130027. |
| 24 | Maia LB, Fonseca L, Moura I, et al. Reduction of carbon dioxide by a molybdenum-containing formate dehydrogenase: a kinetic and mechanistic study [J]. J Am Chem Soc, 2016, 138(28): 8834-8846. |
| 59 | Radon C, Mittelstädt G, Duffus BR, et al. Cryo-EM structures reveal intricate Fe-S cluster arrangement and charging in Rhodobacter capsulatus formate dehydrogenase [J]. Nat Commun, 2020, 11(1): 1912. |
| 25 | da Silva SM, Pimentel C, Valente FMA, et al. Tungsten and molybdenum regulation of formate dehydrogenase expression in Desulfovibrio vulgaris Hildenborough [J]. J Bacteriol, 2011, 193(12): 2909-2916. |
| 26 | Niks D, Duvvuru J, Escalona M, et al. Spectroscopic and kinetic properties of the molybdenum-containing, NAD+-dependent formate dehydrogenase from Ralstonia eutropha [J]. J Biol Chem, 2016, 291(3): 1162-1174. |
| 27 | Schuchmann K, Müller V. Direct and reversible hydrogenation of CO2 to formate by a bacterial carbon dioxide reductase [J]. Science, 2013, 342(6164): 1382-1385. |
| 28 | Bassegoda A, Madden C, Wakerley DW, et al. Reversible interconversion of CO2 and formate by a molybdenum-containing formate dehydrogenase [J]. J Am Chem Soc, 2014, 136(44): 15473-15476. |
| 29 | Mervan Çakar M, Mangas-Sanchez J, Birmingham WR, et al. Discovery of a new metal and NAD+-dependent formate dehydrogenase from Clostridium ljungdahlii [J]. Prep Biochem Biotechnol, 2018, 48(4): 327-334. |
| 30 | Moon M, Park GW, Lee JP, et al. Recombinant expression and characterization of formate dehydrogenase from Clostridium ljungdahlii (ClFDH) as CO2 reductase for converting CO2 to formate [J]. J CO2 Util, 2022, 57: 101876. |
| 31 | Min K, Moon M, Park GW, et al. Newly explored formate dehydrogenases from Clostridium species catalyze carbon dioxide to formate [J]. Bioresour Technol, 2022, 348: 126832. |
| 32 | Alissandratos A, Kim HK, Matthews H, et al. Clostridium carboxidivorans strain P7T recombinant formate dehydrogenase catalyzes reduction of CO2 to formate [J]. Appl Environ Microbiol, 2013, 79(2): 741-744. |
| 33 | Müller U, Willnow P, Ruschig U, et al. Formate dehydrogenase from Pseudomonas oxalaticus [J]. Eur J Biochem, 1978, 83(2): 485-498. |
| 34 | Ruschig U, Müller U, Willnow P, et al. CO2 reduction to formate by NADH catalysed by formate dehydrogenase from Pseudomonas oxalaticus [J]. Eur J Biochem, 1976, 70(2): 325-330. |
| [1] | ZHANG A-na, HAN Xue, GU Tian-yi, XIN Feng-jiao, WANG Yu-lu. Preparation of Low-phenylalanine Casein by Novel Phenylalanine Ammonia-lyases Derived from Rhodotorula [J]. Biotechnology Bulletin, 2024, 40(8): 309-319. |
| [2] | QIAO Ye, ZHANG Nan, YANG Jian-hua, ZHANG Cui-ying, ZHU Lei-lei. Identification and Enzymatic Characterization of a Sugar Phosphatase [J]. Biotechnology Bulletin, 2024, 40(7): 299-306. |
| [3] | HAN Hui, ZHANG Jian, REN Yu-hong. Molecular Modification of the Short-chain Dehydrogenase Lvchun and Its Application in the Synthesis of Chloromycetin [J]. Biotechnology Bulletin, 2023, 39(4): 81-92. |
| [4] | HAO Jun-yao, MA Fu-qiang, YANG Guang-yu. Functional Analysis of Key Residues in the Active Center of Creatinase from Alcaligenes sp. KS-85 [J]. Biotechnology Bulletin, 2021, 37(3): 75-83. |
| [5] | ZHONG Jian-feng, LI Xing-kui, XU Chong-xin, ZHANG Xiao, LIU Xian-jin. Biological Activity of Anti-idiotypic Single Chain Fragment Variable Antibody Against Cry1B by Site-directed Mutagenesis [J]. Biotechnology Bulletin, 2021, 37(10): 186-195. |
| [6] | SUN Xi-lin, JIANG Zhen-yan, LIU Zhi-yi, DAI Lu, SUN Fei, HUANG Wei. Improvement of Thermal Stability of Ganoderma lucidum Protein LZ-8 by Site-directed Mutation of Amino Acids [J]. Biotechnology Bulletin, 2020, 36(1): 23-28. |
| [7] | QIU Jin, HUANG Huo-qing, YAO Bin, LUO Hui-ying. Improvement of Catalytic Activity of Amylase from Bacillus amyloliquefaciens and Its High Expression in Bacillus subtilis [J]. Biotechnology Bulletin, 2019, 35(9): 134-143. |
| [8] | ZHANG Shan-shan, YAO Xiao-long, WANG Ke, YOU Ya. Degradation of Ethyl Formate by Aeromonas salmonicida subsp.salmonicida [J]. Biotechnology Bulletin, 2019, 35(5): 109-117. |
| [9] | HUA Chen, LI Xin-xin, TU Tao, YANG Hong, LUO Hui-ying, CHEN Jia-ming, YAO Bin, BAI Ying-guo, PENG Shu-chuan. Improving the Thermal Stability of Lactate Oxidase by ETSS [J]. Biotechnology Bulletin, 2018, 34(8): 144-150. |
| [10] | MA Bo, ZHANG Ting-ting, WU Bao-xiang. Identification of Filamentous Fungus Strain DWL-C010 and Thermal Stability Analysis of Its Soluble Pigment [J]. Biotechnology Bulletin, 2018, 34(4): 186-193. |
| [11] | REN Tian-lei, YANG Hai-quan, XU Fei. Directed Evolution of Methyl Parathion Hydrolase Based on the Multi-dimensional Features:Molecular Structure and Bioinformatics [J]. Biotechnology Bulletin, 2018, 34(10): 194-200. |
| [12] | Liu Song, Lu Xinyao, Zhou Jingwen, Du Guocheng, Chen Jian. Research Advance on the Structure, Molecular Modification, and Fermentation of Lipoxygenases [J]. Biotechnology Bulletin, 2015, 31(12): 34-41. |
| [13] | Li Tianming, Zhu Linghuan, Liu Tianjia, Dai Baoxin, Feng Huiyong. Synthesis,Expression and Directed Evolution of the Formate Dehydrogenase Gene from Pseudomonas sp. 101 by Saturation Mutagenesis [J]. Biotechnology Bulletin, 2013, 0(5): 105-110. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||