








Biotechnology Bulletin ›› 2025, Vol. 41 ›› Issue (12): 114-123.doi: 10.13560/j.cnki.biotech.bull.1985.2025-0404
Previous Articles Next Articles
GUO Hao-jie( ), WANG Cheng, YANG Fu-rong, DU Bing, MENG Chao-min(
), WANG Cheng, YANG Fu-rong, DU Bing, MENG Chao-min( )
)
Received:2025-04-17
Online:2025-12-26
Published:2026-01-06
Contact:
MENG Chao-min
E-mail:2214442069@qq.com;chaominm@haust.edu.cn
GUO Hao-jie, WANG Cheng, YANG Fu-rong, DU Bing, MENG Chao-min. Cloning and Low-phosphorus Tolerance Analysis of SiSPX9 Gene in Foxtail Millet[J]. Biotechnology Bulletin, 2025, 41(12): 114-123.
| 引物名称 Primer name | 引物序列 Primer sequence (5′-3′) | 引物用途 Primer usage |
|---|---|---|
| SiSPX9-F | ATGAAGTTCGGCAAGCGG | SiSPX9 CDS扩增 |
| SiSPX9-R | TCACTGCGTGGGGATGAG | |
| SiSPX9-qF | TTCGTGGACTACTGCCTGCTC | SiSPX9 RT-qPCR 分析 |
| SiSPX9-qR | GCTTGTCTGCTGCTGCTGTG | |
| Actin-qF | GGCAAACAGGGAGAAGATGA | 谷子内参基因 RT-qPCR 分析 |
| Actin-qR | GAGGTTGTCGGTAAGGTCACG | |
| SiSPX9-pBI121-F | CGCGGATCCATGAAGTTCGGCAAGCGG | pBI121-SiSPX9 载体构建 |
| SiSPX9-pBI121-R | CGCGAGCTCTCACTGCGTGGGGATGAG | |
| 35S-R | GAGCTCCTTTTCCAGCGGACC | 转基因拟南芥分子鉴定 |
Table 1 Primers used in this experiment
| 引物名称 Primer name | 引物序列 Primer sequence (5′-3′) | 引物用途 Primer usage |
|---|---|---|
| SiSPX9-F | ATGAAGTTCGGCAAGCGG | SiSPX9 CDS扩增 |
| SiSPX9-R | TCACTGCGTGGGGATGAG | |
| SiSPX9-qF | TTCGTGGACTACTGCCTGCTC | SiSPX9 RT-qPCR 分析 |
| SiSPX9-qR | GCTTGTCTGCTGCTGCTGTG | |
| Actin-qF | GGCAAACAGGGAGAAGATGA | 谷子内参基因 RT-qPCR 分析 |
| Actin-qR | GAGGTTGTCGGTAAGGTCACG | |
| SiSPX9-pBI121-F | CGCGGATCCATGAAGTTCGGCAAGCGG | pBI121-SiSPX9 载体构建 |
| SiSPX9-pBI121-R | CGCGAGCTCTCACTGCGTGGGGATGAG | |
| 35S-R | GAGCTCCTTTTCCAGCGGACC | 转基因拟南芥分子鉴定 |
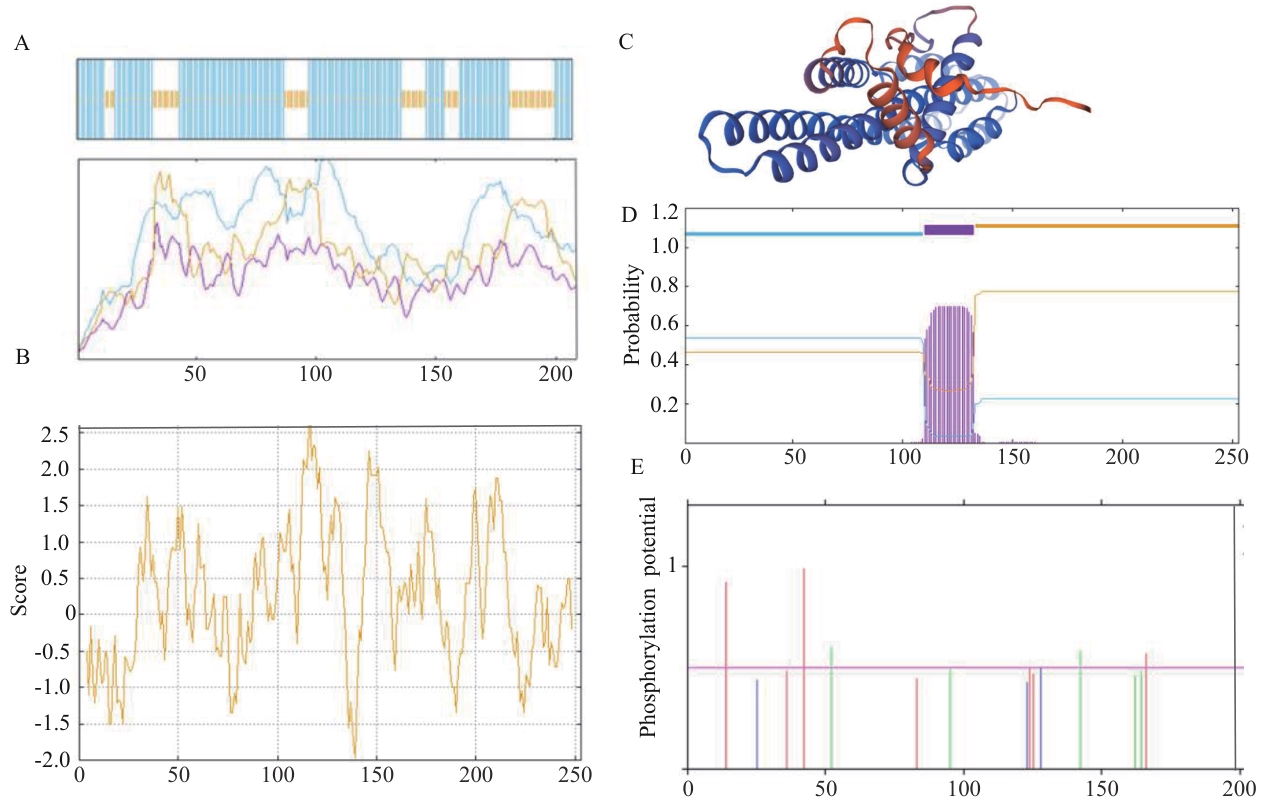
Fig. 3 Bioinformatics analysis ofSiSPX9proteinA: Prediction of secondary structure. B: Tertiary structure of SiSPX9 protein. C: Protein hydrophilicity/hydrophobicity prediction. D: Protein transmembrane structure prediction. E: Phosphorylation site prediction
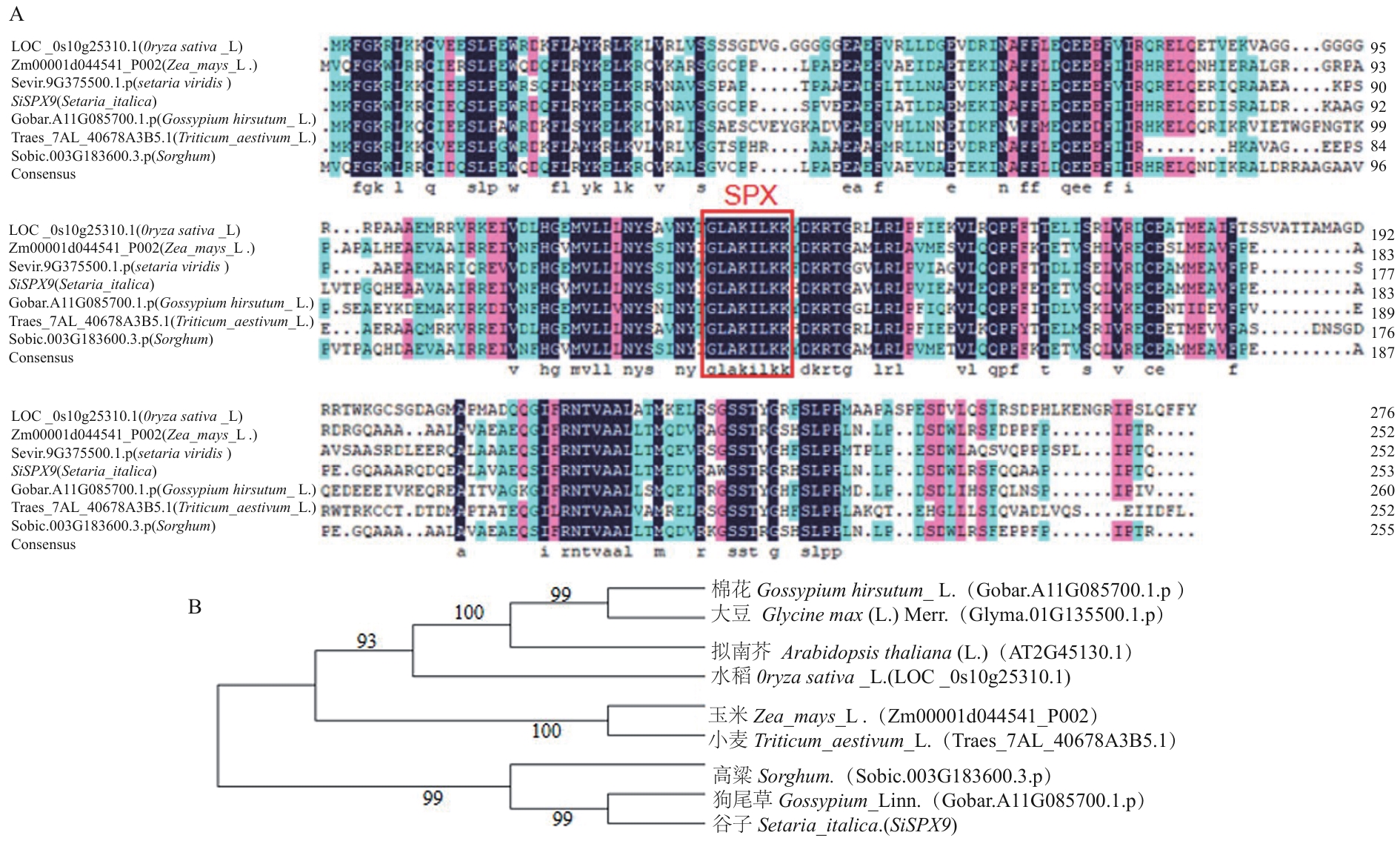
Fig. 4 SiSPX9 protein sequence alignment and phylogenetic tree constructionA: Alignment of SiSPX9protein sequences from different plants. The red box in the diagram contains the domains ofSPX protein. B: Phylogenetic tree analysis of SiSPX9 gene
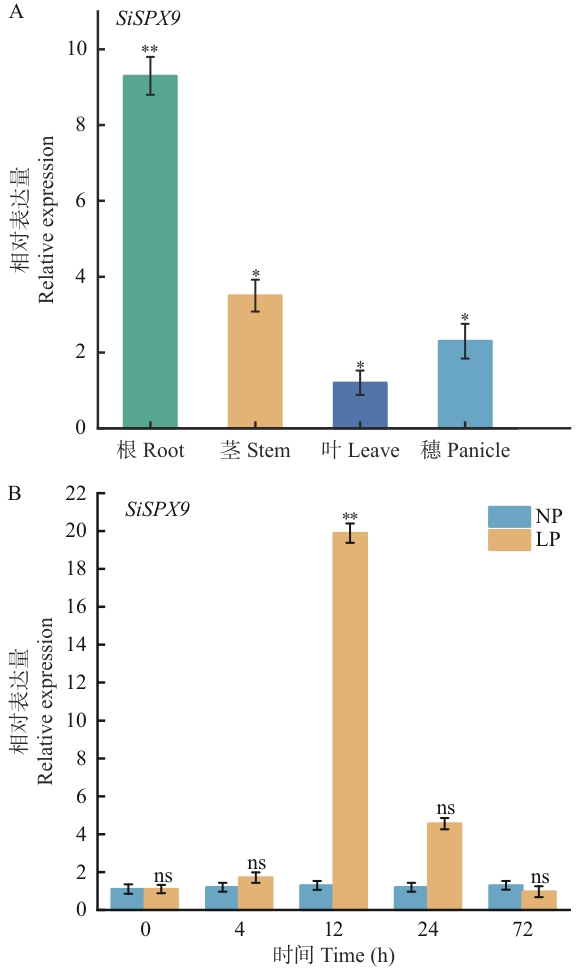
Fig. 5 Tissue-specific and low-phosphorus stress-induced expression of SiSPX9 in foxtail milletA: Expressions of SiSPX9 in different tissues of foxtail millet (Setaria italica). B: Expression patterns of SiSPX9 in roots under low-phosphorus stress at different time points. NP: Normal phosphorus treatment (1 mmol/L KH₂PO₄); LP: Low phosphorus treatment (1 μmol/L KH₂PO₄). *P<0.05, **P< 0.01, ns indicates no significant difference (P>0.05). The same below
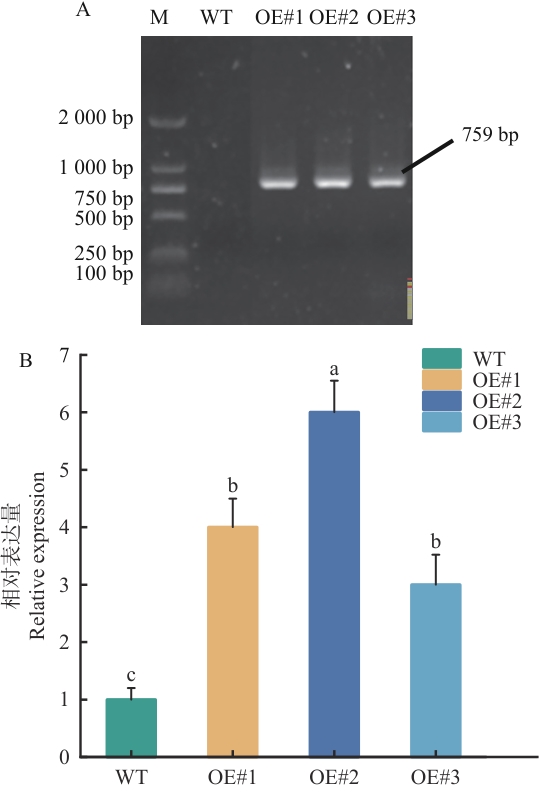
Fig. 6 Identificatied results of transgenic SiSPX9A. thaliana andexpression analysisA: PCR detection of transgenic Arabidopsis lines (M: DL2000 DNA marker; WT: wild-type Arabidopsis; OE#1-OE#3: Arabidopsis transformed with pBI121-SiSPX9). B: qPCR analysis of transgenic Arabidopsis lines. OE#1-OE#3 indicates overexpression-positive lines. Significant differences were determined by Duncan’s new multiple range test (P<0.05). Different lowercase letters indicate statistically significant differences. Error bars indicate the standard deviation (SD) of three independent biological replicates. Relative expression of the target gene is normalized to Actin. The same below
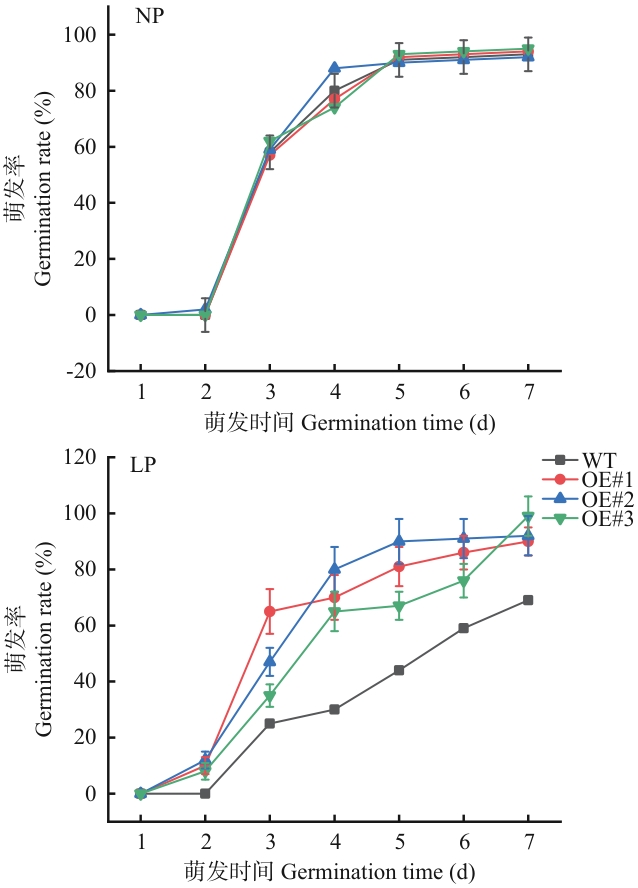
Fig. 7 Germination rates of SiSPX9 transgenic linesThree biological replicates were used for data quantification for each measurement, with 50 seeds per replicate. Each data point indicates the mean of three independent experiments (± standard deviation)
| [1] | 吕建珍, 任莹, 王宏勇, 等. 264份谷子主要育成品种(系)表型多样性综合评价 [J]. 作物杂志, 2022(4): 22-31. |
| Lü JZ, Ren Y, Wang HY, et al. Comprehensive phenotype evaluation of 264 major foxtail millet bred varieties (lines) [J]. Crops, 2022(4): 22-31. | |
| [2] | 李坤杰. 谷子LBD基因家族分析与SiLBD21功能鉴定 [D]. 杨凌: 西北农林科技大学, 2023. |
| Li KJ. Family analysis of LBD gene in millet and functional identification of SiLBD21 [D]. Yangling: Northwest A & F University, 2023. | |
| [3] | 相吉山, 徐峰, 索良喜, 等. 东北地区谷子地方品种和育成品种表型比较分析 [J]. 植物遗传资源学报, 2018, 19(4): 642-656. |
| Xiang JS, Xu F, Suo LX, et al. Comparison on the phenotypic traits between landraces and cultivars of foxtail millet [Setaria italica (L.) P. beauv.] in Northeast China [J]. J Plant Genet Resour, 2018, 19(4): 642-656. | |
| [4] | 许仙菊, 张永春.植物耐低磷胁迫的根系适应性机制研究进展 [J]. 江苏农业学报, 2018, 34(6): 1425-1429. |
| Xu XJ, & Zhang YC. Research advances in root adaptive mechanisms of plant tolerance to low phosphorus stress [J]. Jiangsu J Agric Sci, 2018, 34(6): 1425-1429. | |
| [5] | 黄沆, 付崇允, 周德贵, 等.植物磷吸收的分子机理研究进展 [J]. 分子植物育种, 2008, (1): 117-122. |
| Huang H, Fu CY, Zhou DG, et al. Research advances in molecular mechanisms of plant phosphorus uptake [J]. Mol Plant Breed, 2008 6(1), 117-122. | |
| [6] | Xu WY, Tang WS, Wang CX, et al. SiMYB56 confers drought stress tolerance in transgenic rice by regulating lignin biosynthesis and ABA signaling pathway [J]. Front Plant Sci, 2020, 11: 785. |
| [7] | 吴梦洁, 洪家都, 李芳燕, 等. 闽楠SPX基因家族的鉴定及其缺磷胁迫下的表达分析 [J]. 农业生物技术学报, 2023, 31(9): 1832-1845. |
| Wu MJ, Hong JD, Li FY, et al. Identification of SPX gene family in Phoebe bournei and its expression analysis under phosphorus deficiency stress [J]. J Agric Biotechnol, 2023, 31(9): 1832-1845. | |
| [8] | Jung JY, Ried MK, Hothorn M, et al. Control of plant phosphate homeostasis by inositol pyrophosphates and the SPX domain [J]. Curr Opin Biotechnol, 2018, 49: 156-162. |
| [9] | 晋敏姗, 曲瑞芳, 李红英, 等. 谷子糖转运蛋白基因SiSTPs的鉴定及其参与谷子抗逆胁迫响应的研究 [J]. 作物学报, 2022, 48(4): 825-839. |
| Jin MS, Qu RF, Li HY, et al. Identification of sugar transporter gene family SiSTPs in foxtail millet and its participation in stress response [J]. Acta Agron Sin, 2022, 48(4): 825-839. | |
| [10] | Puga MI, Mateos I, Charukesi R, et al. SPX1 is a phosphate-dependent inhibitor of Phosphate Starvation Response 1 in Arabidopsis [J]. Proc Natl Acad Sci USA, 2014, 111(41): 14947-14952. |
| [11] | 李艳, 方宇辉, 王永霞, 等.不同磷胁迫处理转OsPHR2小麦的转录组学分析 [J]. 作物学报, 2024, 50(2): 340-353. |
| Li Y, Fang YH, Wang YX, et al. Transcriptome analysis of OsPHR2-transgenic wheat under different phosphorus stress treatments [J]. Acta Agron. Sin., 2024, 50(2), 340-353. | |
| [12] | Xiao JB, Xie XM, X, Li C, et al.Identification of SPX family genes in the maize genome and their expression under different phosphate regimes [J]. Plant Biotechnol J, 2021, 168211-220. |
| [13] | 韩长栋. 小麦TaSPX家族鉴定及耐低磷相关基因TaPHO1;H2的功能分析 [D]. 郑州: 河南农业大学, 2023. |
| Han CD. Identification of the TaSPX family in wheat and functional analysis of the low-phosphorus tolerance-related gene TaPHO1;H2 [D]. Zhengzhou: Henan Agricultural University, 2023. | |
| [14] | 商文艳. 小麦SPX基因家族鉴定及TaSPX3在磷胁迫调控中的功能分析 [D]. 郑州: 河南农业大学, 2018. |
| Shang WY. Identification of the wheat SPX gene family and functional analysis of TaSPX3 in phosphorus stress regulation [D]. Zhengzhou: Henan Agricultural University, 2018. | |
| [15] | Wang ZY, Ruan WY, Shi J, et al. Rice SPX1 and SPX2 inhibit phosphate starvation responses through interacting with PHR2 in a phosphate-dependent manner [J]. Proc Natl Acad Sci USA, 2014, 111(41): 14953-14958. |
| [16] | Bustos R, Castrillo G, Linhares F, et al. A central regulatory system largely controls transcriptional activation and repression responses to phosphate starvation in Arabidopsis [J]. PLoS Genet, 2010, 6(9): e1001102. |
| [17] | Plaxton WC, Tran HT. Metabolic adaptations of phosphate-starved plants [J]. Plant Physiol, 2011, 156(3): 1006-1015. |
| [18] | Wang C, Yue WH, Ying YH, et al. Rice SPX-Major Facility Superfamily3, a vacuolar phosphate efflux transporter, is involved in maintaining phosphate homeostasis in rice [J]. Plant Physiol, 2015, 169(4): 2822-2831. |
| [19] | Chen JJ, Han XJ, Liu LX, et al. Genome-wide detection of SPX family and profiling of CoSPX-MFS3 in regulating low-phosphate stress in tea-oil Camellia [J]. Int J Mol Sci, 2023, 24(14): 11552. |
| [20] | Luo JL, Liu ZH, Yan JW, et al. Genome-wide identification of SPX family genes and functional characterization of PeSPX6 and PeSPX-MFS2 in response to low phosphorus in Phyllostachys edulis [J]. Plants, 2023, 12(7): 1496. |
| [21] | Duan K, Yi KK, Dang L, et al. Characterization of a sub-family of Arabidopsis genes with the SPX domain reveals their diverse functions in plant tolerance to phosphorus starvation [J]. Plant J, 2008, 54(6): 965-975. |
| [22] | Pipercevic J, Kohl B, Gerasimaite R, et al. Inositol pyrophosphates activate the vacuolar transport chaperone complex in yeast by disrupting a homotypic SPX domain interaction [J]. Nat Commun, 2023, 14(1): 2645. |
| [23] | Thapa N, Chen M, Horn HT, et al. Phosphatidylinositol-3-OH kinase signalling is spatially organized at endosomal compartments by microtubule-associated protein 4 [J]. Nat Cell Biol, 2020, 22(11): 1357-1370. |
| [24] | 孔晶晶. 玉米ZmLRR1基因调控根系发育的功能和作用机制研究 [D]. 合肥: 安徽农业大学, 2019. |
| Kong JJ. Function and mechanism analysis of ZmLRR1 gene in control of root development in maize [D]. Hefei: Anhui Agricultural University, 2019. | |
| [25] | 李敏. 木薯扩展蛋白家族基因鉴定和功能初步分析 [D]. 海口: 海南大学, 2020. |
| Li M. Genome-wide identification and preliminary functional analysis of expansin gene family in Cassava (Manihot esculenta Crantz) [D]. Haikou: Hainan University, 2020. | |
| [26] | Péret B, Clément M, Nussaume L, et al. Root developmental adaptation to phosphate starvation: better safe than sorry [J]. Trends Plant Sci, 2011, 16(8): 442-450. |
| [27] | 吴敏, 陈瑞, 李贵, 等. 南酸枣苗期耐低磷家系筛选及磷效率评价 [J]. 中南林业科技大学学报, 2025, 45(3): 107-117. |
| Wu M, Chen R, Li G, et al. Screening of low phosphorus tolerant families and phosphorus efficiency of Choerospondias axillaris at seedling stage [J]. J Cent South Univ For Technol, 2025, 45(3): 107-117. | |
| [28] | Dong JS, Piñeros MA, Li XX, et al. An Arabidopsis ABC transporter mediates phosphate deficiency-induced remodeling of root architecture by modulating iron homeostasis in roots [J]. Mol Plant, 2017, 10(2): 244-259. |
| [29] | 戴森焕. PHO1介导磷酸盐转运在植物开花调控中的作用机理 [D]. 杭州: 杭州师范大学, 2024. |
| Dai SH. Mechanism of PHO1-mediated phosphate transport in the regulation of plant flowering [D]. Hangzhou: Hangzhou Normal University, 2024. | |
| [30] | 宗伊琳, 彭枫, 袁满, 等. 植物PHT基因家族功能与磷调控网络研究进展 [J]. 上海师范大学学报: 自然科学版, 2024, 53(6): 751-762. |
| Zong YL, Peng F, Yuan M, et al. Research progress on the functions of plant PHT gene family and phosphorus regulatory network [J]. J Shanghai Norm Univ Nat Sci, 2024, 53(6): 751-762. | |
| [31] | Rodríguez MC, Wawrzyńska A, Sirko A. Intronic T-DNA insertion in Arabidopsis NBR1 conditionally affects wild-type transcript level [J]. Plant Signal Behav, 2014, 9(12): e975659. |
| [1] | CHENG Ting-ting, LIU Jun, WANG Li-li, LIAN Cong-long, WEI Wen-jun, GUO Hui, WU Yao-lin, YANG Jing-fan, LAN Jin-xu, CHEN Sui-qing. Genome-wide Identification of the Chalcone Isomerase Gene Family in Eucommia ulmoides and Analysis of Their Expression Patterns [J]. Biotechnology Bulletin, 2025, 41(9): 242-255. |
| [2] | XU Xiao-ping, YANG Cheng-long, HE Xing, GUO Wen-jie, WU Jian, FANG Shao-zhong. Cloning of the LoAPS1 and Its Function Analysis during the Process of Dormancy Release in Lilium [J]. Biotechnology Bulletin, 2025, 41(9): 195-206. |
| [3] | DONG Xiang-xiang, MIAO Bai-ling, XU He-juan, CHEN Juan-juan, LI Liang-jie, GONG Shou-fu, ZHU Qing-song. Bioinformatics Analysis and Flowering Regulation Function of FveBBX20 Gene in Woodland Strawberry [J]. Biotechnology Bulletin, 2025, 41(9): 115-123. |
| [4] | LI Shan, MA Deng-hui, MA Hong-yi, YAO Wen-kong, YIN Xiao. Identification and Expression Analysis of SKP1 Gene Family in Grapevine (Vitis vinifera L.) [J]. Biotechnology Bulletin, 2025, 41(9): 147-158. |
| [5] | HUANG Guo-dong, DENG Yu-xing, CHENG Hong-wei, DAN Yan-nan, ZHOU Hui-wen, WU Lan-hua. Genome-wide Identification and Expression Analysis of the ZIP Gene Family in Soybean [J]. Biotechnology Bulletin, 2025, 41(9): 71-81. |
| [6] | GONG Hui-ling, XING Yu-jie, MA Jun-xian, CAI Xia, FENG Zai-ping. Identification of Laccase (LAC) Gene Family in Potato (Solanum tuberosum L.) and Its Expression Analysis under Salt Stresses [J]. Biotechnology Bulletin, 2025, 41(9): 82-93. |
| [7] | LIU Jia-li, SONG Jing-rong, ZHAO Wen-yu, ZHANG Xin-yuan, ZHAO Zi-yang, CAO Yi-bo, ZHANG Ling-yun. Identification of the R2R3-MYB Gene and Expression Analysis of Flavonoid Regulatory Genes in Blueberry [J]. Biotechnology Bulletin, 2025, 41(9): 124-138. |
| [8] | ZHANG Yong-yan, GUO Si-jian, LI Jing, HAO Si-yi, LI Rui-de, LIU Jia-peng, CHENG Chun-zhen. Gene Cloning and Functional Analysis of the Anthocyanin-related VcGSTF19 Gene in Blueberry (Vaccinium corymbosum L.) [J]. Biotechnology Bulletin, 2025, 41(9): 139-146. |
| [9] | LAI Shi-yu, LIANG Qiao-lan, WEI Lie-xin, NIU Er-bo, CHEN Ying-e, ZHOU Xin, YANG Si-zheng, WANG Bo. The Role of NbJAZ3 in the Infection of Nicotiana benthamiana by Alfalfa Mosaic Virus [J]. Biotechnology Bulletin, 2025, 41(8): 186-196. |
| [10] | KANG Qin, WANG Xia, SHEN Ming-yang, XU Jing-tian, CHEN Shi-lan, LIAO Ping-yang, XU Wen-zhi, WU Wei, XU Dong-bei. Cloning and Expression Analysis of the UV-B Receptor Gene McUVR8 in Mentha canadensis L. [J]. Biotechnology Bulletin, 2025, 41(8): 255-266. |
| [11] | LA Gui-xiao, ZHAO Yu-long, DAI Dan-dan, YU Yong-liang, GUO Hong-xia, SHI Gui-xia, JIA Hui, YANG Tie-gang. Identification of Plasma Membrane H+-ATPase Gene Family in Safflower and Expression Analysis in Response to Low Nitrogen and Low Phosphorus Stress [J]. Biotechnology Bulletin, 2025, 41(8): 220-233. |
| [12] | LI Kai-jie, WU Yao, LI Dan-dan. Cloning of Gene CtbHLH128 in Safflower and Response Function Regulating Drought Stress [J]. Biotechnology Bulletin, 2025, 41(8): 234-241. |
| [13] | LI Kai-yue, DENG Xiao-xia, YIN Yuan, DU Ya-tong, XU Yuan-jing, WANG Jing-hong, YU Song, LIN Ji-xiang. Identification of LEA Gene Family and Analysis on Its Response to Aluminum Stress in Ricinus communis L. [J]. Biotechnology Bulletin, 2025, 41(7): 128-138. |
| [14] | GONG Yu-han, CHEN Lan, SHANGFANG Hui-zi, HAO Ling-ying, LIU Shuo-qian. Identification and Expression Profile Analysis of the TRB Gene Family in Tea Plant [J]. Biotechnology Bulletin, 2025, 41(7): 214-225. |
| [15] | WEI Yu-jia, LI Yan, KANG Yu-han, GONG Xiao-nan, DU Min, TU Lan, SHI Peng, YU Zi-han, SUN Yan, ZHANG Kun. Cloning and Expression Analysis of the CrMYB4 Gene in Carex rigescens [J]. Biotechnology Bulletin, 2025, 41(7): 248-260. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||