








Biotechnology Bulletin ›› 2025, Vol. 41 ›› Issue (12): 225-239.doi: 10.13560/j.cnki.biotech.bull.1985.2025-0416
Previous Articles Next Articles
ZHANG Xiao-dan1, YIN Zheng1( ), LIU Qing-chen1, LI Xue-mei2, LIU Xiao-hua1(
), LIU Qing-chen1, LI Xue-mei2, LIU Xiao-hua1( ), LIANG Mei-xia1(
), LIANG Mei-xia1( )
)
Received:2025-04-20
Online:2025-12-26
Published:2026-01-06
Contact:
LIU Xiao-hua, LIANG Mei-xia
E-mail:1491091164@qq.com;3305@ldu.edu.cn;mxliangdd@163.com
ZHANG Xiao-dan, YIN Zheng, LIU Qing-chen, LI Xue-mei, LIU Xiao-hua, LIANG Mei-xia. Integrated Metabolomic and Transcriptomic Analysis Reveals the Mechanism of Prunus davidiana Response to Freezing Stress[J]. Biotechnology Bulletin, 2025, 41(12): 225-239.

Fig. 1 Analysis of phenotypic (A) and physiological indices (B, C) of P. davidiana under freezing temperature stressData are means ± standard deviation of three independent experiments. Different letters indicate the significance of differences between groups as determined by one-way analysis of variance (ANOVA) and Duncan’s multiple comparison test (P<0.05). The same below

Fig. 2 Analysis of differentially accumulated metabolites in P. davidiana under different freezing stressesA: Principal component analysis (PCA) of the identified metabolites at different temperatures. B: Heat maps of all differential metabolites (DEMs) at four different temperature treatments. Colours indicate the relative content level of each differential metabolite, with red indicating high content and green indicating low content. C: Volcano plots of CK vs T5, CK vs T15 and CK vs T25 differential metabolites. D: Cluster analysis of all differential metabolites according to the trend of changes, and the detailed clustering results are shown in Figure S2. E: Venn diagrams of CK vs T5, CK vs T15 and CK vs T25 differential metabolites. F: Heat maps of a total of 109 differential metabolites in the three groups of CK vs T5, CK vs T15 and CK vs T25, and the detailed results of differential metabolites are shown in Table S5

Fig. 3 KEGG pathway enrichment analysis of differentially expressed metabolites (DEMs)A-C: KEGG pathway enrichment analysis of the comparisons CK vs T5, CK vs T15, and CK vs T25. D-F: Top 10 differentially expressed metabolites in the comparing CK vs T5, CK vs T15, and CK vs T25
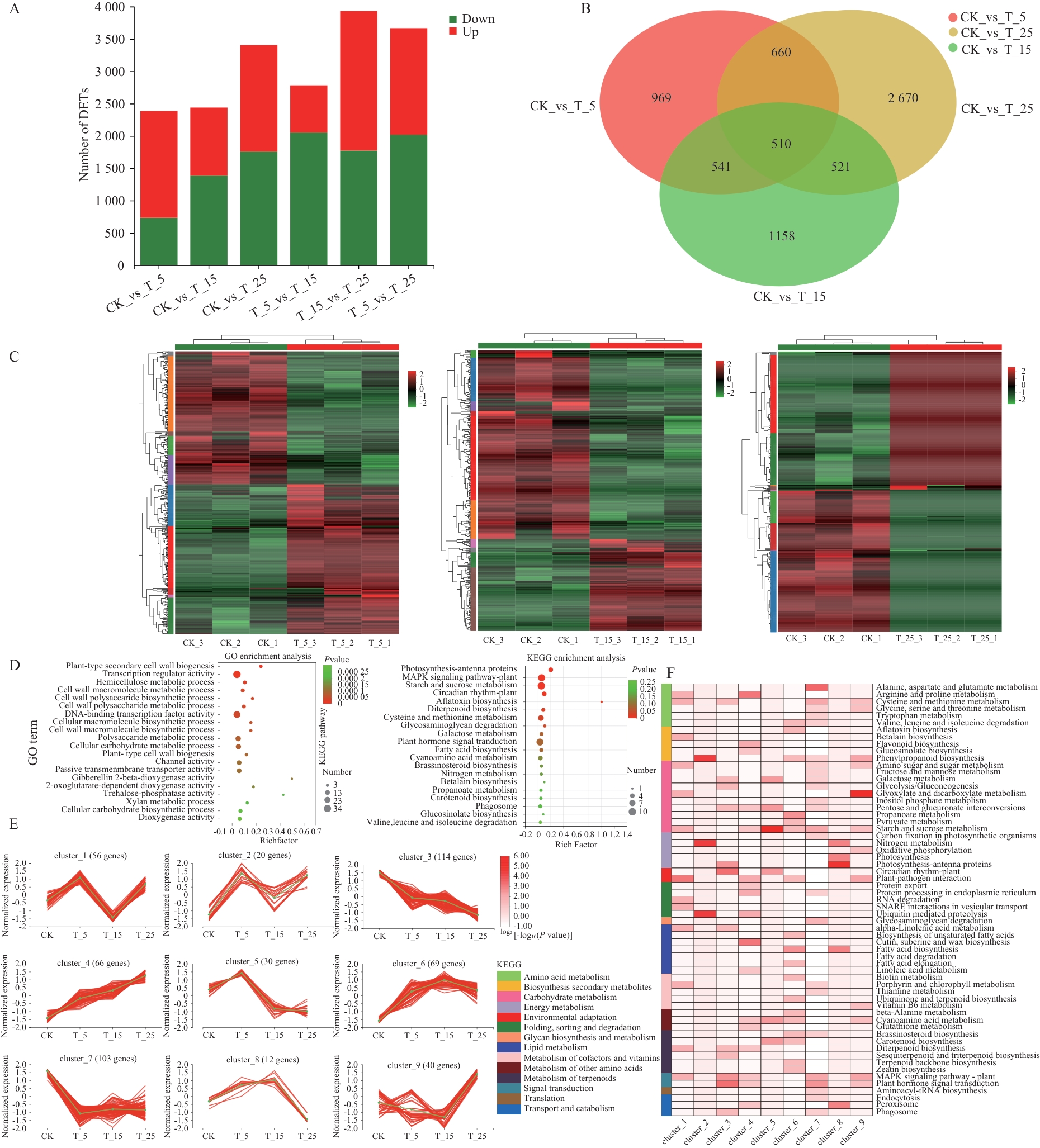
Fig. 4 Expression patterns of differentially expressed genes (DEGs) and GO and KEGG enrichment analyses of co-expressed genesA: Up- and down-regulation of DEGs. B: Venn map of DEGs expression among different treatment groups. C: Co-regulatory relationships of DEGs in all comparison groups. D: Results of KEGG and GO enrichment analyses of top 20 KEGGs and GOs for the 510 co-expressed genes. E: The 510 co-expressed DEGs were classified into 9 clusters. The number of genes in each cluster is shown at the top, the green line indicates the average of the relative expression in each subcluster, and the pink line indicates the relative expressions of individual genes in each subcluster. F: KEGG pathways significantly enriched in each cluster (P<0.05)
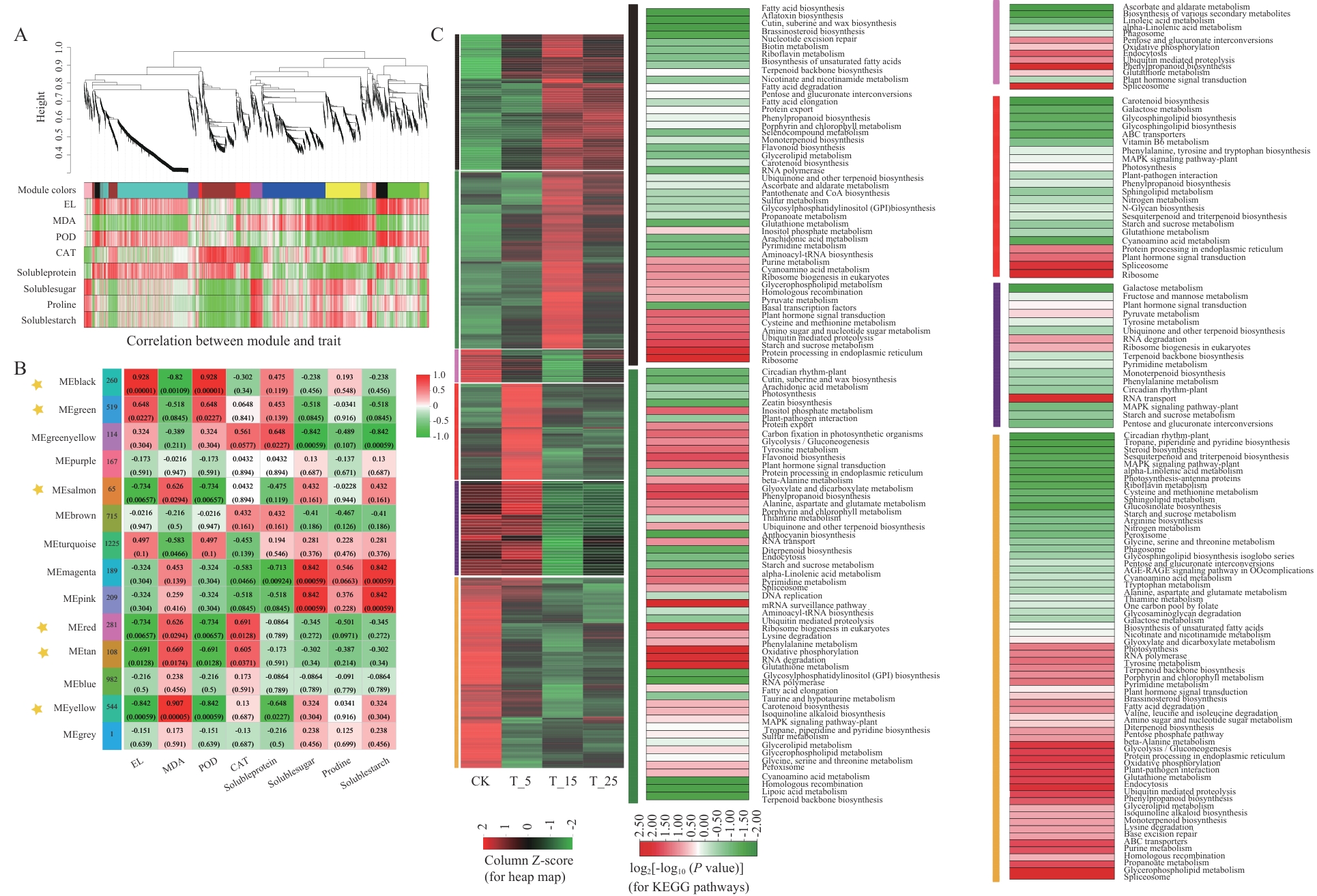
Fig. 6 Association of gene WGCNA analysis with physiological traitsA: Hierarchical clustering tree analysis based on WGCNA, with the main branch divided into 14 modules of different colours. B: Heatmap of module-trait correlation, each row corresponds to one module (colours are consistent with Figure A), and the colours at the intersections of the rows and columns reflect the correlation between the modules and the physiological traits, the black, green, salmon red, red, brownish-brown and yellow modules labelled with an asterisk are significantly (P<0.05) correlated with the electrolyte leakage rate (EL), malondialdehyde (MDA) and peroxidase activity (POD) are significantly correlated (P<0.05). C: Temperature response trend and KEGG pathway enrichment analysis of DEGs in the above six modules (achieved by R language pHYPER function, P<0.05)
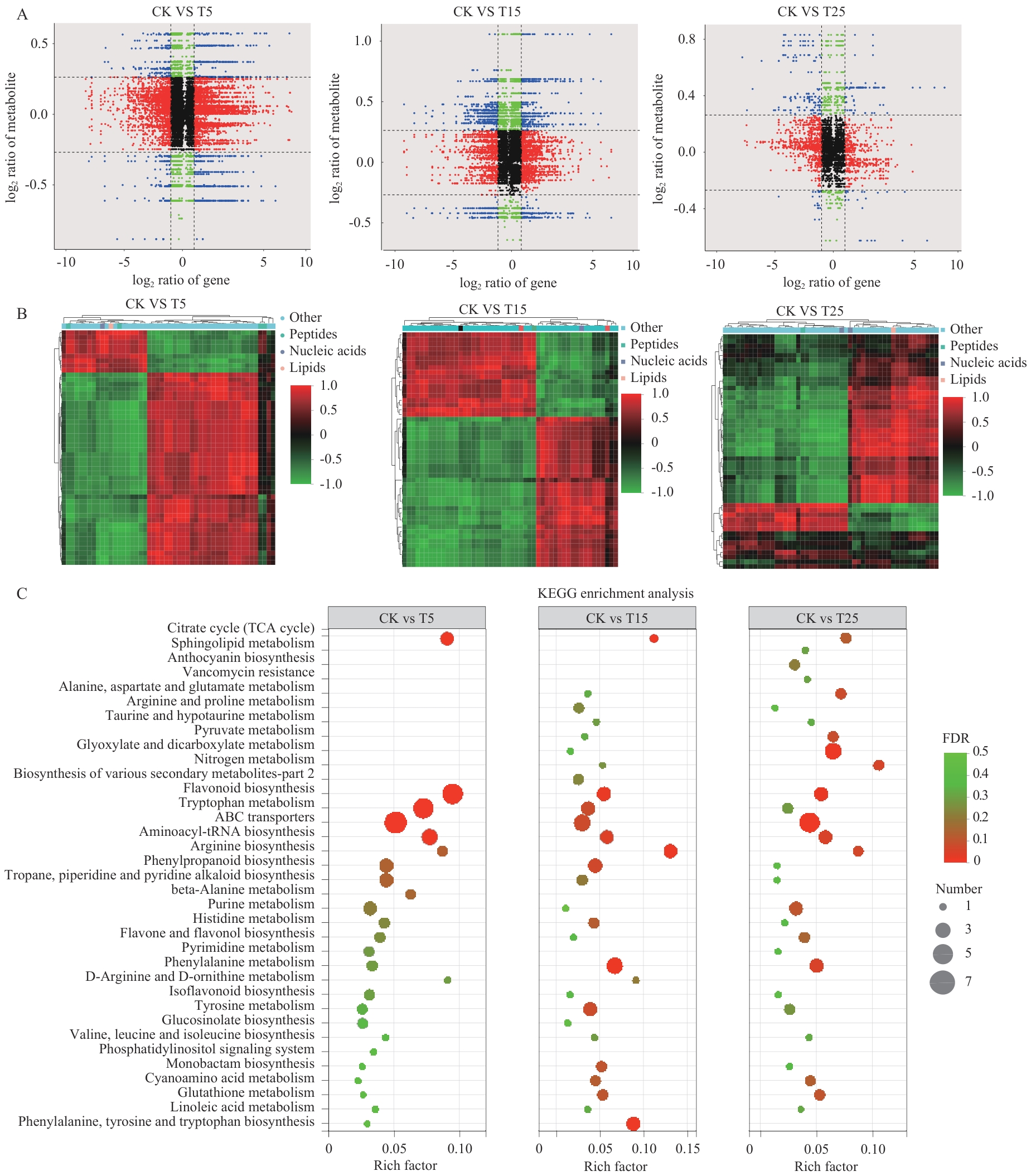
Fig. 7 Metabolome-transcriptome association analysis of P. davidiana in response to different intensities of freezing stressA: Nine-quadrant plot demonstrates metabolite-gene associations, red indicates that both genes and metabolites are upregulated, green indicates that both genes and metabolites are downregulated, black indicates a statistically insignificant association, and blue indicates that the expression trends of genes and metabolites are opposite. B: Clustered heatmap of correlation coefficients of DAMs (PCCs>0.8) between treatment (CK vs T5/T15/T25) and control groups. C: Top 20 DEMs in the comparison of the three groups CK vs T5/T15/T25 KEGG-enriched pathway summary analysis

Fig. 8 Metabolic pathways involved in differentially expressed metabolites (DEMs) and differentially expressed genes (DEGs) in P. davidiana under freezing stressA: Starch and sugar metabolism. B: Organic acid metabolism. C: Flavonoid synthesis. Circles indicate regulated metabolites (green down-regulation, red up-regulation), rectangles indicate regulated genes (green down-regulation, red up-regulation)
| [1] | Wang K, Yin XR, Zhang B, et al. Transcriptomic and metabolic analyses provide new insights into chilling injury in peach fruit [J]. Plant Cell Environ, 2017, 40(8): 1531-1551. |
| [2] | Ding YL, Shi YT, Yang SH. Regulatory networks underlying plant responses and adaptation to cold stress [J]. Annu Rev Genet, 2024, 58(1): 43-65. |
| [3] | Yu QH, Zheng QL, Liu C, et al. Phosphorylation-dependent VaMYB4a regulates cold stress in grapevine by inhibiting VaPIF3 and activating VaCBF4 [J]. Plant Physiol, 2025, 197(2): kiaf035. |
| [4] | Liu XY, Bulley SM, Varkonyi-Gasic E, et al. Kiwifruit bZIP transcription factor AcePosF21 elicits ascorbic acid biosynthesis during cold stress [J]. Plant Physiol, 2023, 192(2): 982-999. |
| [5] | Souza AG, Smiderle OJ, Menegatti RD, et al. Patents for the physiological quality in seeds of peach rootstock classified by weight and stored for different periods [J]. Recent Pat Food Nutr Agric, 2019, 10(2): 124-130. |
| [6] | Javed Tareen M, Wang XK, Ali I, et al. Influence of Scion/Rootstock reciprocal effects on post-harvest and metabolomics regulation in stored peaches [J]. Saudi J Biol Sci, 2022, 29(1): 427-435. |
| [7] | Cao K, Peng Z, Zhao X, et al. Chromosome-level genome assemblies of four wild peach species provide insights into genome evolution and genetic basis of stress resistance [J]. BMC Biol, 2022, 20(1): 139. |
| [8] | Guo ML, Pan RJ, Chu ZJ, et al. Low-temperature stress response: a transcriptomic study of the WRKY family in Prunus davidiana [J]. Cryobiology, 2025, 119: 105252. |
| [9] | Niu RX, Zhao XM, Wang CB, et al. Transcriptome profiling of Prunus persica branches reveals candidate genes potentially involved in freezing tolerance [J]. Sci Hortic, 2020, 259: 108775. |
| [10] | Li YH, Tian QH, Wang ZY, et al. Integrated analysis of transcriptomics and metabolomics of peach under cold stress [J]. Front Plant Sci, 2023, 14: 1153902. |
| [11] | Muthuramalingam P, Shin H, Adarshan S, et al. Molecular insights into freezing stress in peach based on multi-omics and biotechnology: an overview [J]. Plants, 2022, 11(6): 812. |
| [12] | Wang WB, Kim YH, Lee HS, et al. Analysis of antioxidant enzyme activity during germination of alfalfa under salt and drought stresses [J]. Plant Physiol Biochem, 2009, 47(7): 570-577. |
| [13] | Nguyen NH, Van Le B. A simple, economical, and high efficient protocol to produce in vitro miniature rose [J]. Vitro Cell Dev Biol Plant, 2020, 56(3): 362-365. |
| [14] | Wen B, Mei ZL, Zeng CW, et al. metaX: a flexible and comprehensive software for processing metabolomics data [J]. BMC Bioinformatics, 2017, 18(1): 183. |
| [15] | Maruyama K, Urano K, Yoshiwara K, et al. Integrated analysis of the effects of cold and dehydration on rice metabolites, phytohormones, and gene transcripts [J]. Plant Physiol, 2014, 164(4): 1759-1771. |
| [16] | MAN Sambe, He XY, Tu QH, et al. A cold-induced myo-inositol transporter-like gene confers tolerance to multiple abiotic stresses in transgenic tobacco plants [J]. Physiol Plant, 2015, 153(3): 355-364. |
| [17] | Negrão S, Schmöckel SM, Tester M. Evaluating physiological responses of plants to salinity stress [J]. Ann Bot, 2017, 119(1): 1-11. |
| [18] | Liu B, Zhao FM, Cao Y, et al. Photoprotection contributes to freezing tolerance as revealed by RNA-seq profiling of Rhododendron leaves during cold acclimation and deacclimation over time [J]. Hortic Res, 2022, 9: uhab025. |
| [19] | Hao XY, Wang B, Wang L, et al. Comprehensive transcriptome analysis reveals common and specific genes and pathways involved in cold acclimation and cold stress in tea plant leaves [J]. Sci Hortic, 2018, 240: 354-368. |
| [20] | Lu WJ, Wei GQ, Zhou BW, et al. A comparative analysis of photosynthetic function and reactive oxygen species metabolism responses in two Hibiscus cultivars under saline conditions [J]. Plant Physiol Biochem, 2022, 184: 87-97. |
| [21] | Mellado-Mojica E, Calvo-Gómez O, Jofre-Garfias AE, et al. Fructooligosaccharides as molecular markers of geographic origin, growing region, genetic background and prebiotic potential in strawberries: a TLC, HPAEC-PAD and FTIR study [J]. Food Chem Adv, 2022, 1: 100064. |
| [22] | Yoon J, Cho LH, Tun W, et al. Sucrose signaling in higher plants [J]. Plant Sci, 2021, 302: 110703. |
| [23] | Gupta AK, Kaur N. Sugar signalling and gene expression in relation to carbohydrate metabolism under abiotic stresses in plants [J]. J Biosci, 2005, 30(5): 761-776. |
| [24] | Fait A, Fromm H, Walter D, et al. Highway or byway: the metabolic role of the GABA shunt in plants [J]. Trends Plant Sci, 2008, 13(1): 14-19. |
| [25] | Liang X, Wang Y, Shen WX, et al. Genomic and metabolomic insights into the selection and differentiation of bioactive compounds in Citrus [J]. Mol Plant, 2024, 17(11): 1753-1772. |
| [26] | Wang YL, Tong W, Li FD, et al. LUX ARRHYTHMO links CBF pathway and jasmonic acid metabolism to regulate cold tolerance of tea plants [J]. Plant Physiol, 2024, 196(2): 961-978. |
| [27] | Amjadi E, Ganjeali A, Shakeri A, et al. Ecophysiological and phytochemical insights in to Silybum marianum: geographic variations in antioxidant activity and silybin content [J]. Russ J Plant Physiol, 2025, 72(2): 45. |
| [28] | Yang M, Yang J, Su L, et al. Metabolic profile analysis and identification of key metabolites during rice seed germination under low-temperature stress [J]. Plant Sci, 2019, 289: 110282. |
| [29] | Winkel-Shirley B. Flavonoid biosynthesis. a colorful model for genetics, biochemistry, cell biology, and biotechnology [J]. Plant Physiol, 2001, 126(2): 485-493. |
| [30] | Dai H, Xiao CN, Liu HB, et al. Combined NMR and LC-MS analysis reveals the metabonomic changes in Salvia miltiorrhiza Bunge induced by water depletion [J]. J Proteome Res, 2010, 9(3): 1460-1475. |
| [31] | 杨朝结, 张兰, 陈红, 等. 苦荞转录因子基因FtbHLH3调控类黄酮生物合成的功能鉴定 [J]. 生物技术通报, 2025, 41(4): 134-144. |
| Yang CJ, Zhang L, Chen H, et al. Functional identification of the transcription factor gene FtbHLH3 in regulating flavonoid biosynthesis in Fagopyrum tataricum [J]. Biotechnol Bull, 2025, 41(4): 134-144. | |
| [32] | Holton TA, Cornish EC. Genetics and biochemistry of anthocyanin biosynthesis [J]. Plant Cell, 1995, 7(7): 1071-1083. |
| [1] | LIU Yu-shi, LI Zhen, ZOU Yu-chen, TANG Wei-wei, LI Bin. Advances in Spatial Metabolomics in Medicinal Plants [J]. Biotechnology Bulletin, 2025, 41(9): 22-31. |
| [2] | LIU Jian-guo, LIU Ge-er, GUO Ying-xin, WANG Bin, WANG Yu-kun, LU Jin-feng, HUANG Wen-ting, ZHU Yun-na. Integrate Transcriptomic and Metabolomic Analysis of Fruits Quality Differences between ‘Guiyou No. 1’ and ‘Shatianyou’ Pomelo (Citrus maxima) [J]. Biotechnology Bulletin, 2025, 41(9): 168-181. |
| [3] | ZHANG Ya-qi, WANG Qin-qin, SHEN Xia, LI Xu-miao, GAO Min, LI Jun, LI Chen, WANG Hui. Metabolite Early Warning Model for the Risk of Early Progression in Esophageal Squamous Cell Carcinoma [J]. Biotechnology Bulletin, 2025, 41(9): 335-344. |
| [4] | JIANG Tian-wei, MA Pei-jie, LI Ya-jiao, CHEN Cai-jun, LIU Xiao-xia, WANG Xiao-li. Metabolic Response Analysis of Brachypodium distachyon to Photoperiods [J]. Biotechnology Bulletin, 2025, 41(7): 237-247. |
| [5] | YANG Zong-hui, LI Li-bin, MENG Zhao-juan, GAO Tian, ZHU Li-xia, DU Hai-mei, DONG Wei-wei, CAO Qi-wei. Comparative Transcriptomics Reveals Synergistic Control of Cucumber Sex Determination by Ethylene Signaling and Epigenetic Regulation [J]. Biotechnology Bulletin, 2025, 41(12): 139-155. |
| [6] | LUO Yi-fei, XU Ya, LU Si-qi, GAO Yuan-yuan, GOU Zhan, FANG Hong, SHANG Yi, TENG Lin-lin. Analysis of Volatile Components in Solanum tuberosum L. Plants Infected with Phytophthora infestans [J]. Biotechnology Bulletin, 2025, 41(11): 247-260. |
| [7] | WANG Rui, QI Ji. Integrating Histological Image Information to Enhance Cell Clustering Resolution in Spatial Transcriptome [J]. Biotechnology Bulletin, 2024, 40(8): 39-46. |
| [8] | XU Pei-dong, YI Jian-feng, CHEN Di, CHEN Hao, XIE Bing-yan, ZHAO Wen-jun. Progress in the Application of Omics Technology in Biocontrol Bacillus [J]. Biotechnology Bulletin, 2024, 40(10): 208-220. |
| [9] | HAN Le-le, SONG Wen-di, BIAN Jia-shen, LI Yang, YANG Shuang-sheng, CHEN Zi-yi, LI Xiao-wei. Revealing the Flavonoid Biosynthesis of Soybean GmERD15c under Salt Stress by Combined Analysis of Transcriptome and Metabolome [J]. Biotechnology Bulletin, 2024, 40(10): 243-252. |
| [10] | JIANG Yu-shan, LAN Qian, WANG Fang, JIANG Liang, PEI Cheng-cheng. Characterization of a Quinoa Mutant Affecting Tyrosine Metabolism [J]. Biotechnology Bulletin, 2024, 40(10): 253-261. |
| [11] | ZHAO Rui, DI Jing-yi, ZHANG Guang-tong, LIU Hao, GAO Wei-xia. Screening Endogenous Expression Elements in Streptococcus zooepidemicus via Transcriptomics Analysis and Applications for High Yield of Hyaluronic Acid [J]. Biotechnology Bulletin, 2024, 40(10): 296-304. |
| [12] | HE Shi-yu, ZENG Zhong-da, LI Bo-yan. Application Progress of Spatially Resolved Metabolomics in Disease Diagnosis Research [J]. Biotechnology Bulletin, 2024, 40(1): 145-159. |
| [13] | ZHOU Ai-ting, PENG Rui-qi, WANG Fang, WU Jian-rong, MA Huan-cheng. Analysis of Metabolic Differences of Biocontrol Strain DZY6715 at Different Growth Stages [J]. Biotechnology Bulletin, 2023, 39(9): 225-235. |
| [14] | HAN Hua-rui, YANG Yu-lu, MEN Yi-han, HAN Shang-ling, HAN Yuan-huai, HUO Yi-qiong, HOU Si-yu. SiYABBYs Involved in Rhamnoside Biosynthesis During the Flower Development of Setaria italica, Based on Metabolomics [J]. Biotechnology Bulletin, 2023, 39(6): 189-198. |
| [15] | XU Yang, DING Hong, ZHANG Guan-chu, GUO Qing, ZHANG Zhi-meng, DAI Liang-xiang. Metabolomics Analysis of Germinating Peanut Seed Under Salt Stress [J]. Biotechnology Bulletin, 2023, 39(1): 199-213. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||