








Biotechnology Bulletin ›› 2024, Vol. 40 ›› Issue (10): 296-304.doi: 10.13560/j.cnki.biotech.bull.1985.2024-0541
Previous Articles Next Articles
ZHAO Rui( ), DI Jing-yi, ZHANG Guang-tong, LIU Hao, GAO Wei-xia(
), DI Jing-yi, ZHANG Guang-tong, LIU Hao, GAO Wei-xia( )
)
Received:2024-06-07
Online:2024-10-26
Published:2024-11-20
Contact:
GAO Wei-xia
E-mail:zr1375649659@163.com;gaoweixia@tust.edu.cn
ZHAO Rui, DI Jing-yi, ZHANG Guang-tong, LIU Hao, GAO Wei-xia. Screening Endogenous Expression Elements in Streptococcus zooepidemicus via Transcriptomics Analysis and Applications for High Yield of Hyaluronic Acid[J]. Biotechnology Bulletin, 2024, 40(10): 296-304.
| 表达元件Expression element | 表达元件控制的基因功能Gene functions controlled by expression elements | FPKM | 大小Size/bp |
|---|---|---|---|
| PR0 | cp25(常见的用于兽疫链球菌的异源表达元件)[ | - | 59 |
| PR1 | 3-phosphoshikimate 1-carboxyvinyltransferase | 65.45 | 130 |
| PR2 | Conserved hypothetical protein | 65.68 | 423 |
| PR3 | tRNA-specific 2-thiouridylase MnmA | 65.97 | 283 |
| PR4 | Maltodextrin transport system permease protein MalF | 89.14 | 299 |
| PR5 | Alkaline phosphatase synthesis transcriptional regulatory protein PhoP | 97.38 | 149 |
| PR6 | Sugar uptake protein | 105.48 | 500 |
| PR7 | 16S rRNA m(2)G 1207 methyltransferase | 117.68 | 233 |
| PR8 | Glutamate dehydrogenase | 137.6 | 500 |
| PR9 | UDP-N-acetylmuramoyl-L-alanyl-D-glutamate synthetase | 430.41 | 152 |
| PR10 | Putative cell-cycle regulation histidine triad protein HIT | 436.44 | 472 |
| PR11 | L-lactate dehydrogenase | 561.89 | 191 |
| PR12 | ABC transporter ATP-binding protein | 644.25 | 120 |
| PR13 | S-adenosylmethionine:tRNA ribosyltransferase-isomerase | 708.09 | 268 |
| PR14 | Methylated-DNA--protein-cysteine methyltransferase | 709.89 | 492 |
| PR15 | 30S ribosomal protein S4 | 855.87 | 500 |
| PR16 | 50S ribosomal protein L28 | 1029.4 | 500 |
| PR17 | Cell division protein | 1152.84 | 211 |
| PR18 | 50S ribosomal protein L32 | 1734.26 | 269 |
| PR19 | NADH oxidase | 1980.71 | 249 |
| PR20 | 30S ribosomal protein S13 | 2550.44 | 159 |
| PR21 | Fructose-bisphosphate aldolase | 4387.94 | 200 |
| PR22 | 6-phosphofructokinase | 4644.17 | 79 |
| PR23 | phosphoglycerate kinase | 4912.63 | 200 |
| PR24 | C3-bisphosphoglycerate-dependent phosphoglycerate mutase GpmA | 4918.12 | 122 |
| PR25 | UDP-glucose 6-dehydrogenase HasB | 6662.7 | 253 |
| PR26 | Hyaluronan synthase HasA | 7891.36 | 396 |
| PR27 | Glyceraldehyde-3-phosphate dehydrogenase | 9036.98 | 112 |
| PR28 | Cold shock protein | 10105.49 | 206 |
| PR29 | Conserved hypothetical protein | 11533.49 | 500 |
| PR30 | Hypothetical protein | 12563.03 | 340 |
| PR31 | Szp protein | 13899.81 | 384 |
| PR32 | Phosphopyruvate hydratase | 20029.66 | 213 |
Table 1 Information about the expression elements in S. zooepidemicus
| 表达元件Expression element | 表达元件控制的基因功能Gene functions controlled by expression elements | FPKM | 大小Size/bp |
|---|---|---|---|
| PR0 | cp25(常见的用于兽疫链球菌的异源表达元件)[ | - | 59 |
| PR1 | 3-phosphoshikimate 1-carboxyvinyltransferase | 65.45 | 130 |
| PR2 | Conserved hypothetical protein | 65.68 | 423 |
| PR3 | tRNA-specific 2-thiouridylase MnmA | 65.97 | 283 |
| PR4 | Maltodextrin transport system permease protein MalF | 89.14 | 299 |
| PR5 | Alkaline phosphatase synthesis transcriptional regulatory protein PhoP | 97.38 | 149 |
| PR6 | Sugar uptake protein | 105.48 | 500 |
| PR7 | 16S rRNA m(2)G 1207 methyltransferase | 117.68 | 233 |
| PR8 | Glutamate dehydrogenase | 137.6 | 500 |
| PR9 | UDP-N-acetylmuramoyl-L-alanyl-D-glutamate synthetase | 430.41 | 152 |
| PR10 | Putative cell-cycle regulation histidine triad protein HIT | 436.44 | 472 |
| PR11 | L-lactate dehydrogenase | 561.89 | 191 |
| PR12 | ABC transporter ATP-binding protein | 644.25 | 120 |
| PR13 | S-adenosylmethionine:tRNA ribosyltransferase-isomerase | 708.09 | 268 |
| PR14 | Methylated-DNA--protein-cysteine methyltransferase | 709.89 | 492 |
| PR15 | 30S ribosomal protein S4 | 855.87 | 500 |
| PR16 | 50S ribosomal protein L28 | 1029.4 | 500 |
| PR17 | Cell division protein | 1152.84 | 211 |
| PR18 | 50S ribosomal protein L32 | 1734.26 | 269 |
| PR19 | NADH oxidase | 1980.71 | 249 |
| PR20 | 30S ribosomal protein S13 | 2550.44 | 159 |
| PR21 | Fructose-bisphosphate aldolase | 4387.94 | 200 |
| PR22 | 6-phosphofructokinase | 4644.17 | 79 |
| PR23 | phosphoglycerate kinase | 4912.63 | 200 |
| PR24 | C3-bisphosphoglycerate-dependent phosphoglycerate mutase GpmA | 4918.12 | 122 |
| PR25 | UDP-glucose 6-dehydrogenase HasB | 6662.7 | 253 |
| PR26 | Hyaluronan synthase HasA | 7891.36 | 396 |
| PR27 | Glyceraldehyde-3-phosphate dehydrogenase | 9036.98 | 112 |
| PR28 | Cold shock protein | 10105.49 | 206 |
| PR29 | Conserved hypothetical protein | 11533.49 | 500 |
| PR30 | Hypothetical protein | 12563.03 | 340 |
| PR31 | Szp protein | 13899.81 | 384 |
| PR32 | Phosphopyruvate hydratase | 20029.66 | 213 |
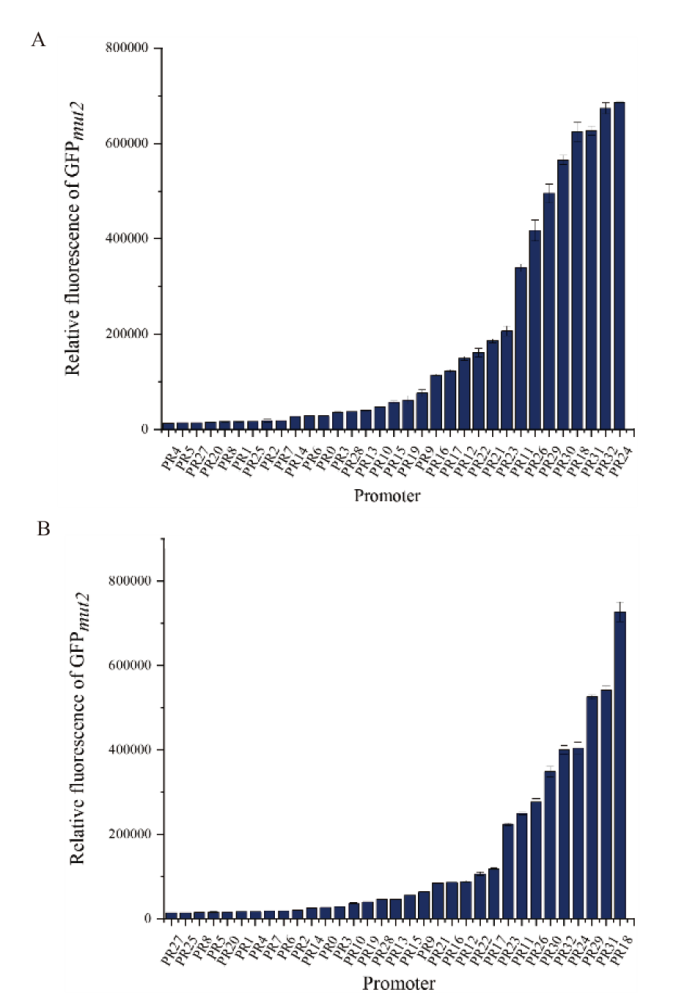
Fig. 2 Relative fluorescence intensity of GFP expressed by different promoter Relative fluorescence intensity of GFP(fluorescence intensity/OD600)cultured in THY medium(A)or FSB medium(B)for 8 h

Fig. 3 Relative transcriptions of gfp gene in different medium and fermentation time A, B: The relative transcriptions of gfp controlled by different expression elements cultured in THY medium for 4 h and 8 h, respectively. C, D: The relative transcriptions of gfp controlled by different expression elements cultured in FSB medium for 4 h and 8 h, respectively
| 培养基 Medium | 组成型强表达元件 Constitutive strongly expressive elements | 指数期强表达元件 Strongly expressed elements in exponential phase | 稳定期强表达元件 Strongly expressed elements in stable phase |
|---|---|---|---|
| THY | PR15、PR17、PR26、PR30、PR32 | PR8、PR11、PR16、PR18、PR19、PR21、PR24、PR29 | PR12、PR31 |
| FSB | PR8、PR16、PR24、PR26、PR28、PR30、PR31 | PR5、PR13 | PR6、PR12、PR21、PR22、PR23、PR32 |
Table 2 Various strong expression elements
| 培养基 Medium | 组成型强表达元件 Constitutive strongly expressive elements | 指数期强表达元件 Strongly expressed elements in exponential phase | 稳定期强表达元件 Strongly expressed elements in stable phase |
|---|---|---|---|
| THY | PR15、PR17、PR26、PR30、PR32 | PR8、PR11、PR16、PR18、PR19、PR21、PR24、PR29 | PR12、PR31 |
| FSB | PR8、PR16、PR24、PR26、PR28、PR30、PR31 | PR5、PR13 | PR6、PR12、PR21、PR22、PR23、PR32 |
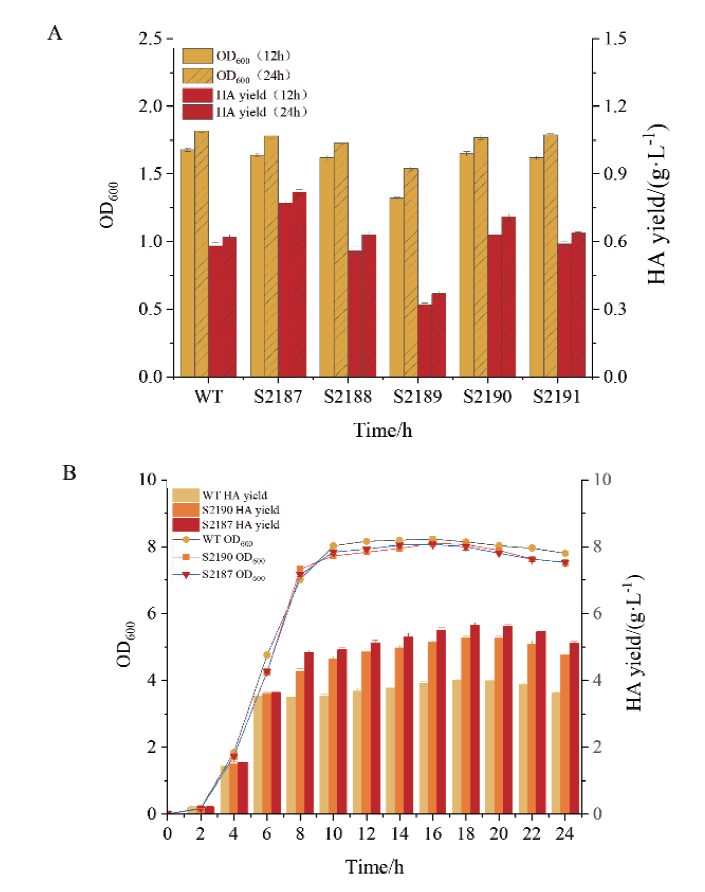
Fig. 5 Growth and HA yield of strains A: Growth and HA yield of hasA, hasB, hasC, hasD, and hasE overexpressing strains shaken at 12 h and 24 h. B: Growth curve and HA yield of S2187 (hasA) and S2190 (hasD) overexpressing strains cultured in 2 L fermenter
| [1] |
Stern R, Asari AA, Sugahara KN. Hyaluronan fragments: an information-rich system[J]. Eur J Cell Biol, 2006, 85(8): 699-715.
doi: 10.1016/j.ejcb.2006.05.009 pmid: 16822580 |
| [2] | Graça MFP, Miguel SP, Cabral CSD, et al. Hyaluronic acid—based wound dressings: a review[J]. Carbohydr Polym, 2020, 241: 116364. |
| [3] | Diane R, Leonardo MS, Bruno F, et al. Hyaluronic acid-from production to application: a review[J]. Biointerface Res Appl Chem, 2022, 13(3): 211-233. |
| [4] | Zheng XL, Wang BT, Tang X, et al. Absorption, metabolism, and functions of hyaluronic acid and its therapeutic prospects in combination with microorganisms: a review[J]. Carbohydr Polym, 2023, 299: 120153. |
| [5] | Johnson ME, Murphy PJ, Boulton M. Effectiveness of sodium hyaluronate eyedrops in the treatment of dry eye[J]. Graefes Arch Clin Exp Ophthalmol, 2006, 244(1): 109-112. |
| [6] | Armstrong DC, Johns MR. Culture conditions affect the molecular weight properties of hyaluronic acid produced by Streptococcus zooepidemicus[J]. Appl Environ Microbiol, 1997, 63(7): 2759-2764. |
| [7] |
Pummill PE, DeAngelis PL. Alteration of polysaccharide size distribution of a vertebrate hyaluronan synthase by mutation[J]. J Biol Chem, 2003, 278(22): 19808-19814.
doi: 10.1074/jbc.M301097200 pmid: 12654925 |
| [8] |
Keasling JD. Manufacturing molecules through metabolic engineering[J]. Science, 2010, 330(6009): 1355-1358.
doi: 10.1126/science.1193990 pmid: 21127247 |
| [9] | Manfrão-Netto JHC, Queiroz EB, de Oliveira Junqueira AC, et al. Genetic strategies for improving hyaluronic acid production in recombinant bacterial culture[J]. J Appl Microbiol, 2022, 132(2): 822-840. |
| [10] |
Liu L, Liu YF, Li JH, et al. Microbial production of hyaluronic acid: current state, challenges, and perspectives[J]. Microb Cell Fact, 2011, 10: 99.
doi: 10.1186/1475-2859-10-99 pmid: 22088095 |
| [11] |
Afrasiabi S, Zanjani FSA, Ahmadian G, et al. The effect of manipulating glucuronic acid biosynthetic pathway in Bacillus subtilis strain on hyaluronic acid production[J]. AMB Express, 2023, 13(1): 63.
doi: 10.1186/s13568-023-01567-2 pmid: 37354246 |
| [12] | Woo JE, Seong HJ, Lee SY, et al. Metabolic engineering of Escher-ichia coli for the production of hyaluronic acid from glucose and galactose[J]. Front Bioeng Biotechnol, 2019, 7: 351. |
| [13] | Gomes AMV, Netto JHCM, Carvalho LS, et al. Heterologous hyaluronic acid production in Kluyveromyces lactis[J]. Microorganisms, 2019, 7(9): 294. |
| [14] | Chien LJ, Lee CK. Hyaluronic acid production by recombinant Lactococcus lactis[J]. Appl Microbiol Biotechnol, 2007, 77(2): 339-346. |
| [15] | Wang Y, Hu LT, Huang H, et al. Eliminating the capsule-like layer to promote glucose uptake for hyaluronan production by engineered Corynebacterium glutamicum[J]. Nat Commun, 2020, 11(1): 3120. |
| [16] |
Chong BF, Blank LM, McLaughlin R, et al. Microbial hyaluronic acid production[J]. Appl Microbiol Biotechnol, 2005, 66(4): 341-351.
doi: 10.1007/s00253-004-1774-4 pmid: 15599518 |
| [17] |
Hynes WL, Walton SL. Hyaluronidases of Gram-positive bacteria[J]. FEMS Microbiol Lett, 2000, 183(2): 201-207.
pmid: 10675584 |
| [18] | Mohan N, Pavan SS, Achar A, et al. Calorespirometric investigation of Streptococcus zooepidemicus metabolism: Thermodynamics of anabolic payload contribution by growth and hyaluronic acid synthesis[J]. Biochem Eng J, 2019, 152: 107367. |
| [19] | Beres SB, Sesso R, Pinto SWL, et al. Genome sequence of a Lancefield Group C Streptococcus zooepidemicus strain causing epidemic nephritis: new information about an old disease[J]. PLoS One, 2008, 3(8): e3026. |
| [20] | Cimini D, Iacono ID, Carlino E, et al. Engineering S. equi subsp. zooepidemicus towards concurrent production of hyaluronic acid and chondroitin biopolymers of biomedical interest[J]. AMB Express, 2017, 7(1): 61. |
| [21] | Chen WY, Marcellin E, Hung J, et al. Hyaluronan molecular weight is controlled by UDP-N-acetylglucosamine concentration in Streptococcus zooepidemicus[J]. J Biol Chem, 2009, 284(27): 18007-18014. |
| [22] | Chen WY, Marcellin E, Steen JA, et al. The role of hyaluronic acid precursor concentrations in molecular weight control in Streptococcus zooepidemicus[J]. Mol Biotechnol, 2014, 56(2): 147-156. |
| [23] | Sun XY, Yang DD, Wang YY, et al. Development of a markerless gene deletion system for Streptococcus zooepidemicus: functional characterization of hyaluronan synthase gene[J]. Appl Microbiol Biotechnol, 2013, 97(19): 8629-8636. |
| [24] | Sun XQ, Wang Z, Bi YL, et al. Genetic and functional characterization of the hyaluronate lyase HylB and the beta-N-acetylglucosaminidase HylZ in Streptococcus zooepidemicus[J]. Curr Microbiol, 2015, 70(1): 35-42. |
| [25] | Gao WX, Xie YY, Zuo M, et al. Improved genetic transformation by disarmament of type II Restriction-Modification system in Streptococcus zooepidemicus[J]. 3 Biotech, 2022, 12(9): 192. |
| [26] | Sullivan MJ, Ulett GC. Stable expression of modified green fluorescent protein in group B streptococci to enable visualization in experimental systems[J]. Appl Environ Microbiol, 2018, 84(18): e01262-18. |
| [27] | Amado IR, Vázquez JA, Pastrana L, et al. Microbial production of hyaluronic acid from agro-industrial by-products: molasses and corn steep liquor[J]. Biochem Eng J, 2017, 117: 181-187. |
| [28] | de Macedo AC, Santana MHA. Hyaluronic acid depolymerization by ascorbate-redox effects on solid state cultivation of Streptococcus zooepidemicus in cashew apple fruit bagasse[J]. World J Microbiol Biotechnol, 2012, 28(5): 2213-2219. |
| [29] | Pires AMB, Macedo AC, Eguchi SY, et al. Microbial production of hyaluronic acid from agricultural resource derivatives[J]. Bioresour Technol, 2010, 101(16): 6506-6509. |
| [30] | Shah MV, Badle SS, Ramachandran KB. Hyaluronic acid production and molecular weight improvement by redirection of carbon flux towards its biosynthesis pathway[J]. Biochem Eng J, 2013, 80: 53-60. |
| [31] | Zhou SH, Ding RP, Chen J, et al. Obtaining a panel of cascade promoter-5'-UTR complexes in Escherichia coli[J]. ACS Synth Biol, 2017, 6(6): 1065-1075. |
| [32] | Song YF, Nikoloff JM, Fu G, et al. Promoter screening from Bacillus subtilis in various conditions hunting for synthetic biology and industrial applications[J]. PLoS One, 2016, 11(7): e0158447. |
| [33] | 刘秀霞, 赵子豪, 孙杨, 等. 谷氨酸棒杆菌内源表达元件的筛选[J]. 微生物学通报, 2016, 43(8): 1671-1678. |
| Liu XX, Zhao ZH, Sun Y, et al. Selection of endogenous expression elements from Corynebactrium glutamicum[J]. Microbiol China, 2016, 43(8): 1671-1678. | |
| [34] | 郑小娥, 王震, 刘浩. 兽疫链球菌 ATCC39920可控诱导表达系统的构建及其应用[J]. 生物技术通报, 2016, 32(12): 130-136. |
| Zheng XE, Wang Z, Liu H. Construction of controlled inducible expression system in Streptococcus zooepidemicus ATCC39920 and its application[J]. Biotechnol Bull, 2016, 32(12): 130-136. | |
| [35] | Chahuki FF, Aminzadeh S, Jafarian V, et al. Hyaluronic acid production enhancement via genetically modification and culture medium optimization in Lactobacillus acidophilus[J]. Int J Biol Macromol, 2019, 121: 870-881. |
| [36] | Jin P, Kang Z, Yuan PH, et al. Production of specific-molecular-weight hyaluronan by metabolically engineered Bacillus subtilis 168[J]. Metab Eng, 2016, 35: 21-30. |
| [1] | WU Shu-ning, SU Yong-ping, LI Dong-xue, BAI Ying-guo, LIU Bo, ZHANG Zhi-wei. Design and Application of a Cumate-inducible Promoter for Corynebacterium glutamicum [J]. Biotechnology Bulletin, 2024, 40(7): 108-116. |
| [2] | LI Meng-ran, YE Wei, LI Sai-ni, ZHANG Wei-yang, LI Jian-jun, ZHANG Wei-min. Expression of Lithocarols Biosynthesis Gene litI and Functional Analysis of Its Promoter [J]. Biotechnology Bulletin, 2024, 40(6): 310-318. |
| [3] | WANG Zhou, YU Jie, WANG Jin-hua, WANG Yong-ze, ZHAO Xiao. Anaerobic Expression of Lactate Dehydrogenase to Improve the D-lactic Acid Optical Purity Procluced by Escherichia coli [J]. Biotechnology Bulletin, 2024, 40(5): 290-299. |
| [4] | ZHANG Qing-lan, ZHANG Ya-ran, JU Zhi-hua, WANG Xiu-ge, XIAO Yao, WANG Jin-peng, WEI Xiao-chao, GAO Ya-ping, BAI Fu-heng, WANG Hong-cheng. Identification and Transcriptional Regulation Analysis of Core Promoter in Bovine TARDBP Gene [J]. Biotechnology Bulletin, 2024, 40(4): 306-318. |
| [5] | ZHANG Jie-ping, GUAN Yue-feng. Crop Breeding Based on Promoter Editing [J]. Biotechnology Bulletin, 2024, 40(10): 53-61. |
| [6] | ZHU Yi, LIU Tang-jing, GONG Guo-yi, ZHANG Jie, WANG Jin-fang, ZHANG Hai-ying. Cloning and Expression Analysis of ClPP2C3 in Citrullus lanatus [J]. Biotechnology Bulletin, 2024, 40(1): 243-249. |
| [7] | LIU Yu-ling, WANG Meng-yao, SUN Qi, MA Li-hua, ZHU Xin-xia. Effect of RD29A Promoter on the Stress Resistance of Transgenic Tobacco with SikCDPK1 Gene from Saussurea involucrata [J]. Biotechnology Bulletin, 2023, 39(9): 168-175. |
| [8] | GUO San-bao, SONG Mei-ling, LI Ling-xin, YAO Zi-zhao, GUI Ming-ming, HUANG Sheng-he. Cloning and Analysis of Chalcone Synthase Gene and Its Promoter from Euphorbia maculata [J]. Biotechnology Bulletin, 2023, 39(4): 148-156. |
| [9] | YANG Lan, ZHANG Chen-xi, FAN Xue-wei, WANG Yang-guang, WANG Chun-xiu, LI Wen-ting. Gene Cloning, Expression Pattern, and Promoter Activity Analysis of Chicken BMP15 [J]. Biotechnology Bulletin, 2023, 39(4): 304-312. |
| [10] | SHI Guang-zhen, WANG Zhao-ye, SUN Qi, ZHU Xin-xia. Cloning and Activity Analysis of SikCDPK1 Promoter from Saussurea involucrata [J]. Biotechnology Bulletin, 2022, 38(9): 191-197. |
| [11] | CHEN Guang, LI Jia, DU Rui-ying, WANG Xu. pOsHAK1:OsFLN2 Expression Enhances the Drought Tolerance by Altering Sugar Metabolism in Rice [J]. Biotechnology Bulletin, 2022, 38(8): 92-100. |
| [12] | NIE Li-bin, YI Ling-xin, DENG Yan, SHENG Qi, WU Xiao-yu, ZHANG Bin. Pathway Engineering Modification of Corynebacterium glutamicum for Shikimic Acid Production [J]. Biotechnology Bulletin, 2022, 38(6): 93-102. |
| [13] | HAO Qing-qing, YAO Sheng, LIU Jia-he, CHEN Pei-zhen, ZHANG Meng-yang, JI Kong-shu. Cloning and Expression Analysis of NAC Transcription Factor PmNAC8 in Pinus massoniana [J]. Biotechnology Bulletin, 2022, 38(4): 202-216. |
| [14] | YE Peng-lin, Kwasi Kyere-Yeboah, GAO E-bin. Effects on the Biosynthesis of Ethanol by Promoters PpetE and Pcpc560 in Synechocystis sp. PCC 6803 [J]. Biotechnology Bulletin, 2022, 38(2): 141-149. |
| [15] | MA Yan-qin, QIU Yi-bin, LI Sha, XU Hong. Research Progress in the Biosynthesis and Metabolic Engineering of Hyaluronic Acid [J]. Biotechnology Bulletin, 2022, 38(2): 252-262. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||