








Biotechnology Bulletin ›› 2025, Vol. 41 ›› Issue (12): 280-293.doi: 10.13560/j.cnki.biotech.bull.1985.2025-0452
Previous Articles Next Articles
SI Xu-peng1,2( ), CUI Xiu-wen1,2, OUYANG Li-zhi3, FANG Dan-dan1,2, PEI Long-ying1,2, FANG Gui-ping1,2, PU Xi-lei1,2, MA Yi-mian3, ZHANG Zheng3(
), CUI Xiu-wen1,2, OUYANG Li-zhi3, FANG Dan-dan1,2, PEI Long-ying1,2, FANG Gui-ping1,2, PU Xi-lei1,2, MA Yi-mian3, ZHANG Zheng3( )
)
Received:2025-05-01
Online:2025-12-26
Published:2026-01-06
Contact:
ZHANG Zheng
E-mail:sixupeng0527@163.com;zhangzheng@implad.ac.cn
SI Xu-peng, CUI Xiu-wen, OUYANG Li-zhi, FANG Dan-dan, PEI Long-ying, FANG Gui-ping, PU Xi-lei, MA Yi-mian, ZHANG Zheng. Transcriptome Analysis of Different Parts of Ferula Sinkiangensis K. M. Shen and Exploration of Genes Related to Sesquiterpene Synthesis[J]. Biotechnology Bulletin, 2025, 41(12): 280-293.
基因名 Gene name | 基因ID Gene ID | 引物序列 Primer sequence (5′-3′) |
|---|---|---|
| P450 | Cluster-8529.5884-F | CGGTCCGAGAAGTTATGGGG |
| Cluster-8529.5884-R | TGGCAACCACCCTCTTCATC | |
| DXS | Cluster-8529.15434-F | GCTGGACATGGGTGTATGCT |
| Cluster-8529.15434-R | CTGGAGTTTTGCTCCCCCAT | |
| AACT | Cluster-8529.15178-F | CTGCGGTTAAATCTGCTGGT |
| Cluster-8529.15178-R | GTTACTCCCAAAGGATGCCCC | |
| TPS | Cluster-8529.5582-F | TACTGGATGATGAGGGAGCGA |
| Cluster-8529.5582-R | ACAATACGAGTGGAGCCGC | |
| HMGR | Cluster-8529.7778-F | TTCGAGACGCTTGCACTCAT |
| Cluster-8529.7778-R | TTGCATCCCCAGTGCTACAG | |
| DXR | Cluster-8529.20686-F | TGTCAGAAGCACCAAGACGA |
| Cluster-8529.20686-R | TACAAGAGCGGGACTCAAGC | |
| Actin | Actin-F | TGGTATTGTGCTGGATTCTGGT |
| Actin-R | TGAGATCACCACCAGCAAGG |
Table 1 Primer sequence for RT-qPCR
基因名 Gene name | 基因ID Gene ID | 引物序列 Primer sequence (5′-3′) |
|---|---|---|
| P450 | Cluster-8529.5884-F | CGGTCCGAGAAGTTATGGGG |
| Cluster-8529.5884-R | TGGCAACCACCCTCTTCATC | |
| DXS | Cluster-8529.15434-F | GCTGGACATGGGTGTATGCT |
| Cluster-8529.15434-R | CTGGAGTTTTGCTCCCCCAT | |
| AACT | Cluster-8529.15178-F | CTGCGGTTAAATCTGCTGGT |
| Cluster-8529.15178-R | GTTACTCCCAAAGGATGCCCC | |
| TPS | Cluster-8529.5582-F | TACTGGATGATGAGGGAGCGA |
| Cluster-8529.5582-R | ACAATACGAGTGGAGCCGC | |
| HMGR | Cluster-8529.7778-F | TTCGAGACGCTTGCACTCAT |
| Cluster-8529.7778-R | TTGCATCCCCAGTGCTACAG | |
| DXR | Cluster-8529.20686-F | TGTCAGAAGCACCAAGACGA |
| Cluster-8529.20686-R | TACAAGAGCGGGACTCAAGC | |
| Actin | Actin-F | TGGTATTGTGCTGGATTCTGGT |
| Actin-R | TGAGATCACCACCAGCAAGG |
| 序号 Serial number | 通路ID Pathway ID | 代谢通路 Metabolic pathway | 数量 Quantity |
|---|---|---|---|
| 1 | ko00523 | Polyketide sugar unit biosynthesis | 3 |
| 2 | ko00900 | Terpenoid backbone biosynthesis | 59 |
| 3 | ko00902 | Monoterpenoid biosynthesis | 7 |
| 4 | ko00903 | Limonene degradation | 12 |
| 5 | ko00904 | Diterpenoid biosynthesis | 24 |
| 6 | ko00905 | Brassinosteroid biosynthesis | 18 |
| 7 | ko00906 | Carotenoid biosynthesis | 33 |
| 8 | ko00908 | Zeatin biosynthesis | 34 |
| 9 | ko00909 | Sesquiterpenoid and triterpenoid biosynthesis | 15 |
| 10 | ko01051 | Biosynthesis of ansamycins | 2 |
| 11 | ko01053 | Biosynthesis of siderophore group nonribosomal peptides | 1 |
Table 2 KEGG pathway analysis of terpenoids and polyketides
| 序号 Serial number | 通路ID Pathway ID | 代谢通路 Metabolic pathway | 数量 Quantity |
|---|---|---|---|
| 1 | ko00523 | Polyketide sugar unit biosynthesis | 3 |
| 2 | ko00900 | Terpenoid backbone biosynthesis | 59 |
| 3 | ko00902 | Monoterpenoid biosynthesis | 7 |
| 4 | ko00903 | Limonene degradation | 12 |
| 5 | ko00904 | Diterpenoid biosynthesis | 24 |
| 6 | ko00905 | Brassinosteroid biosynthesis | 18 |
| 7 | ko00906 | Carotenoid biosynthesis | 33 |
| 8 | ko00908 | Zeatin biosynthesis | 34 |
| 9 | ko00909 | Sesquiterpenoid and triterpenoid biosynthesis | 15 |
| 10 | ko01051 | Biosynthesis of ansamycins | 2 |
| 11 | ko01053 | Biosynthesis of siderophore group nonribosomal peptides | 1 |

Fig. 4 Pathway closely related to the formation of pharmacological substancesA: Biosynthesis of other secondary metabolites. B: Metabolism of terpenoids and polyketides
| 骨架 Skeleton | 名称 Name | 数量 Quantity | FPKM (Fragments Per) |
|---|---|---|---|
萜类骨架 Terpenoid skeleton(59)ko00900 | Geranylgeranyl pyrophosphate synthase | 3 | 6.62-14.69 |
| Farnesol kinase | 1 | 4.79-7.41 | |
| 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase (MDS) | 1 | 5.3-6.11 | |
| 2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase (MCT) | 3 | 10.67-29.75 | |
| Prenyl protein peptidase | 1 | 3.66-6.47 | |
| Diphosphomevalonate decarboxylase (MVD) | 2 | 4.33-10.76 | |
| Hydroxymethylglutaryl-CoA synthase (HMGCS) | 2 | 6.53-37.72 | |
| Farnesylcysteine lyase | 1 | 39-45.25 | |
| Geranylgeranyl diphosphate reductase | 2 | 3.17-46.74 | |
| Acetyl-CoA C-acetyltransferase (ACAT) | 1 | 23.22-41.51 | |
| Polycis-polyprenyl diphosphate synthase | 2 | 2.29-22.71 | |
| Geranyl diphosphate synthase (GPS) | 3 | 0.39-14.17 | |
| 4-Hydroxy-3-methylbut-2-enyl diphosphate reductase (HDR) | 5 | 0.1-25.06 | |
| Hypothetical protein | 1 | 0.38-1.52 | |
| 1-Deoxy-D-xylulose-5-phosphate synthase (DXS) | 3 | 3.58-43.29 | |
| 3-Hydroxy-3-methylglutaryl-coenzyme A reductase (HMGR) | 3 | 0.43-60.75 | |
| 4-Hydroxy-3-methylbut-2-enyl-diphosphate synthase (HXS) | 1 | 25.93-42.74 | |
| CAAX prenyl protease | 1 | 60.08-65.9 | |
| Mevalonate kinase (MK) | 1 | 16.24-18.02 | |
| Phosphomevalonate kinase (MVK2) | 1 | 8.52-11.37 | |
| Isopentenyl-diphosphate Delta-isomerase | 1 | 75.21-102.72 | |
| Acetyl-CoA C-acetyltransferase | 4 | 0.16-83.75 | |
| Protein-S-isoprenylcysteine O-methyltransferase (STE14) | 1 | 10.42-17.69 | |
| Isopentenyl phosphate kinase | 1 | 8.28-10.05 | |
| Farnesyl diphosphate synthase (FDPS) | 1 | 5.67-7.64 | |
| Dihydroflavonol-4-reductase | 3 | 0.42-18.19 | |
| Prenylcysteine alpha-carboxyl methylesterase | 1 | 3.53-4.22 | |
| Protein farnesyltransferase/geranylgeranyltransferase | 1 | 13.1-14.01 | |
| Geranylgeranyl pyrophosphate synthase/Polyprenyl synthetase | 1 | 6.66-8.04 | |
| 4-Diphosphocytidyl-2-C-methyl-D-erythritol kinase (CMK) | 1 | 12.65-15.84 | |
| Solanesyl-diphosphate synthase | 1 | 2.35-3.24 | |
| Protein farnesyltransferase subunit beta (FNTB) | 1 | 8.47-9.76 | |
| Ditrans,polycis-polyprenyl diphosphate synthase | 1 | 11.54-14.64 | |
| Prenyl protease | 1 | 12.7-13.94 | |
倍半萜及三萜生物合成 Biosynthesis of sesquiterpenes and triterpenes(15)ko00909 | Sesquiterpene synthase | 3 | 0.33-10.01 |
| Beta-amyrin synthase | 2 | 1.05-3.53 | |
| Squalene epoxidase | 4 | 0.14-32.06 | |
| Dihydroflavonol-4-reductase | 3 | 0.16-18.19 | |
| Squalene synthase | 2 | 0.42-58.1 | |
| (3S, 6E)-Nerolidol synthase | 1 | 1.97-3.91 |
Table 3 Unigenes related to terpenoid backbone and sesquiterpene synthesis in F. sinkiangensis
| 骨架 Skeleton | 名称 Name | 数量 Quantity | FPKM (Fragments Per) |
|---|---|---|---|
萜类骨架 Terpenoid skeleton(59)ko00900 | Geranylgeranyl pyrophosphate synthase | 3 | 6.62-14.69 |
| Farnesol kinase | 1 | 4.79-7.41 | |
| 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase (MDS) | 1 | 5.3-6.11 | |
| 2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase (MCT) | 3 | 10.67-29.75 | |
| Prenyl protein peptidase | 1 | 3.66-6.47 | |
| Diphosphomevalonate decarboxylase (MVD) | 2 | 4.33-10.76 | |
| Hydroxymethylglutaryl-CoA synthase (HMGCS) | 2 | 6.53-37.72 | |
| Farnesylcysteine lyase | 1 | 39-45.25 | |
| Geranylgeranyl diphosphate reductase | 2 | 3.17-46.74 | |
| Acetyl-CoA C-acetyltransferase (ACAT) | 1 | 23.22-41.51 | |
| Polycis-polyprenyl diphosphate synthase | 2 | 2.29-22.71 | |
| Geranyl diphosphate synthase (GPS) | 3 | 0.39-14.17 | |
| 4-Hydroxy-3-methylbut-2-enyl diphosphate reductase (HDR) | 5 | 0.1-25.06 | |
| Hypothetical protein | 1 | 0.38-1.52 | |
| 1-Deoxy-D-xylulose-5-phosphate synthase (DXS) | 3 | 3.58-43.29 | |
| 3-Hydroxy-3-methylglutaryl-coenzyme A reductase (HMGR) | 3 | 0.43-60.75 | |
| 4-Hydroxy-3-methylbut-2-enyl-diphosphate synthase (HXS) | 1 | 25.93-42.74 | |
| CAAX prenyl protease | 1 | 60.08-65.9 | |
| Mevalonate kinase (MK) | 1 | 16.24-18.02 | |
| Phosphomevalonate kinase (MVK2) | 1 | 8.52-11.37 | |
| Isopentenyl-diphosphate Delta-isomerase | 1 | 75.21-102.72 | |
| Acetyl-CoA C-acetyltransferase | 4 | 0.16-83.75 | |
| Protein-S-isoprenylcysteine O-methyltransferase (STE14) | 1 | 10.42-17.69 | |
| Isopentenyl phosphate kinase | 1 | 8.28-10.05 | |
| Farnesyl diphosphate synthase (FDPS) | 1 | 5.67-7.64 | |
| Dihydroflavonol-4-reductase | 3 | 0.42-18.19 | |
| Prenylcysteine alpha-carboxyl methylesterase | 1 | 3.53-4.22 | |
| Protein farnesyltransferase/geranylgeranyltransferase | 1 | 13.1-14.01 | |
| Geranylgeranyl pyrophosphate synthase/Polyprenyl synthetase | 1 | 6.66-8.04 | |
| 4-Diphosphocytidyl-2-C-methyl-D-erythritol kinase (CMK) | 1 | 12.65-15.84 | |
| Solanesyl-diphosphate synthase | 1 | 2.35-3.24 | |
| Protein farnesyltransferase subunit beta (FNTB) | 1 | 8.47-9.76 | |
| Ditrans,polycis-polyprenyl diphosphate synthase | 1 | 11.54-14.64 | |
| Prenyl protease | 1 | 12.7-13.94 | |
倍半萜及三萜生物合成 Biosynthesis of sesquiterpenes and triterpenes(15)ko00909 | Sesquiterpene synthase | 3 | 0.33-10.01 |
| Beta-amyrin synthase | 2 | 1.05-3.53 | |
| Squalene epoxidase | 4 | 0.14-32.06 | |
| Dihydroflavonol-4-reductase | 3 | 0.16-18.19 | |
| Squalene synthase | 2 | 0.42-58.1 | |
| (3S, 6E)-Nerolidol synthase | 1 | 1.97-3.91 |
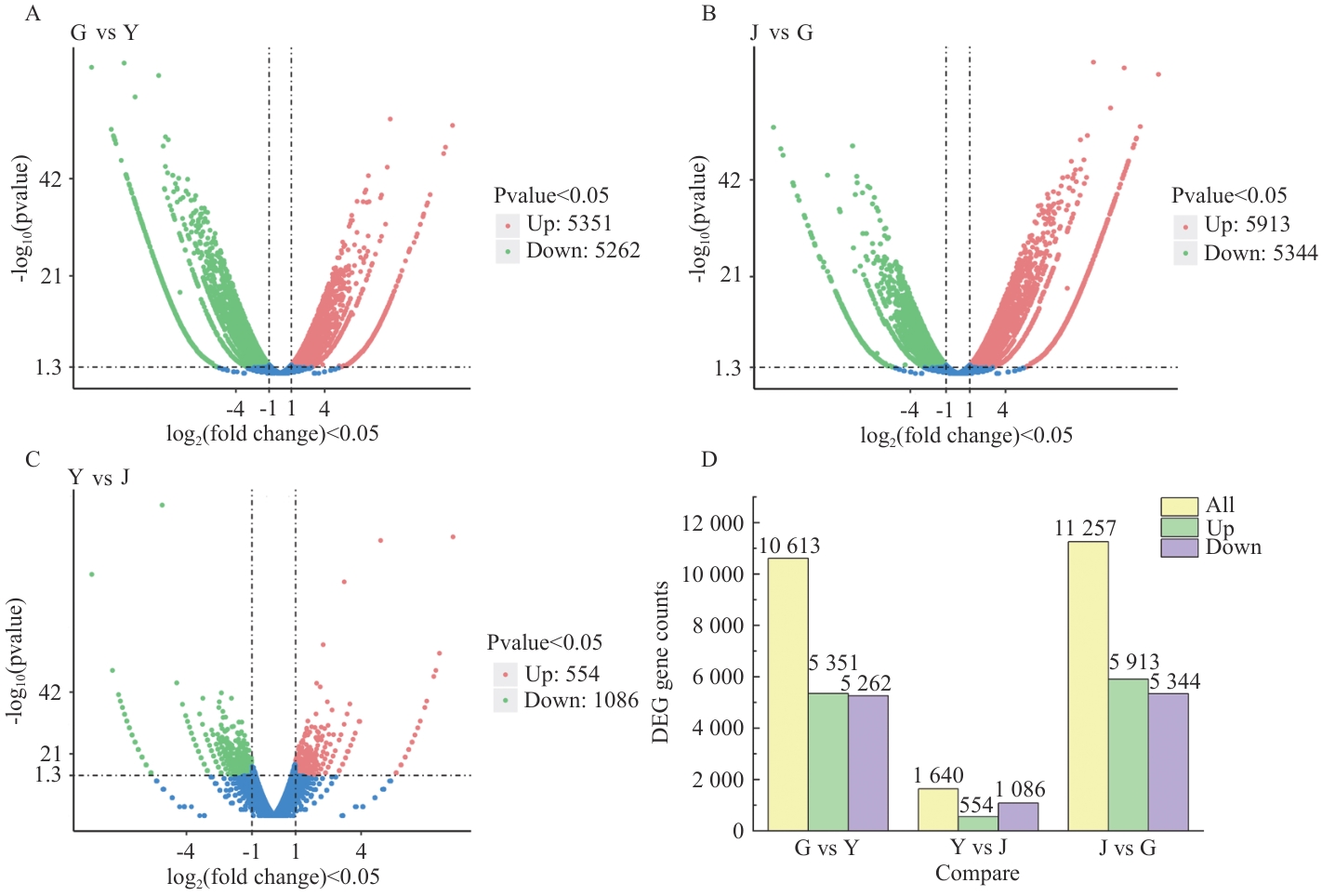
Fig. 5 Distribution and expression of differentially expressed genes in different parts of F. sinkiangensisA-C: Distribution of differentially expressed genes. D: Expression of differential genes
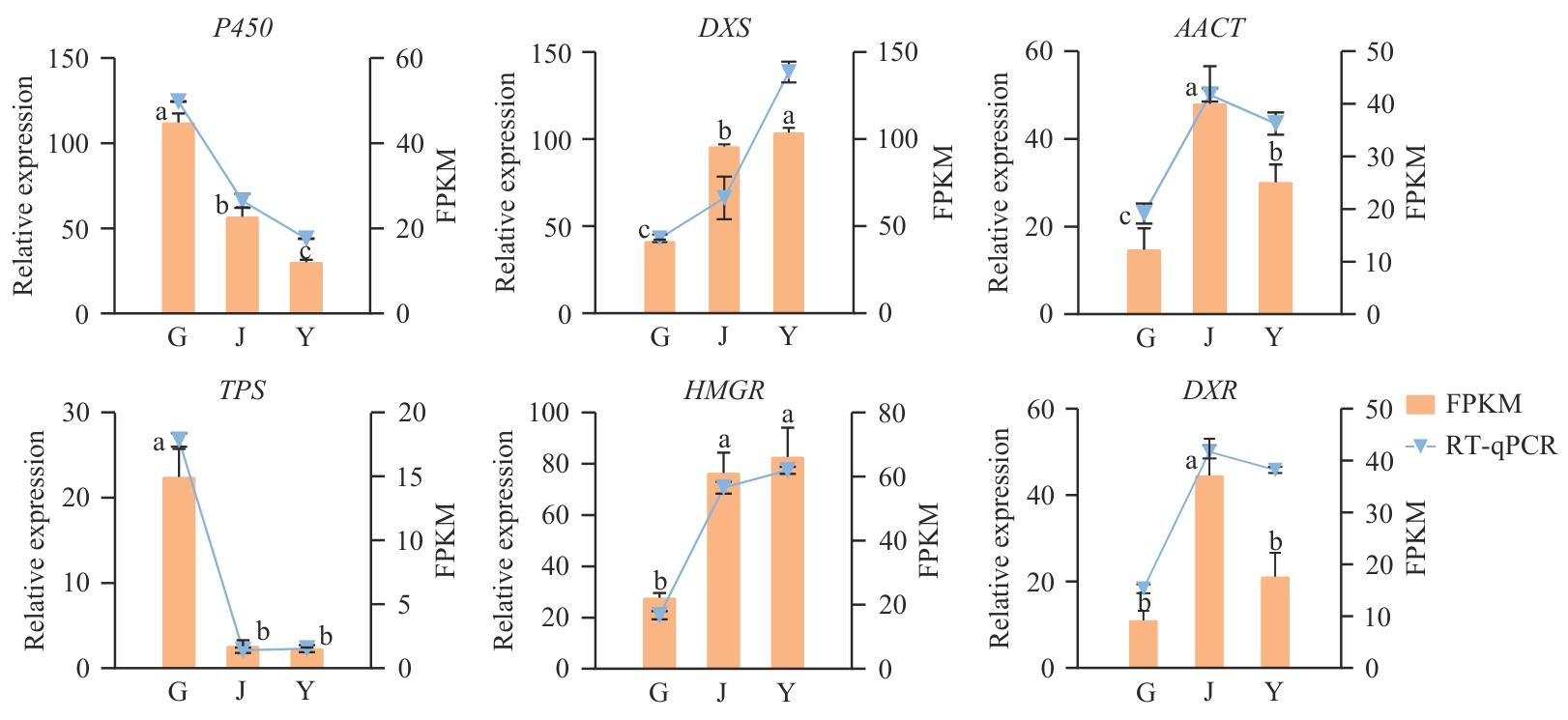
Fig. 9 Verification of gene expressions related to sesquiterpene synthesis in different parts of F. sinkiangensisData with different lowercase letters indicate significant difference at 0.05 level
重复基序数目 Number of repeated motifs | SSR位点数目 Number of SSR loci | 总数量 Total quantity | |||||
|---|---|---|---|---|---|---|---|
单核苷酸 Single nucleotide | 二核苷酸 Dinucleotide | 三核苷酸 Trinucleotide | 四核苷酸 Tetranucleotide | 五核苷酸 Pentanucleotide | 六核苷酸 Hexanucleotide | ||
| 5 | 0 | 0 | 844 | 96 | 27 | 41 | 1 008 |
| 6 | 0 | 1800 | 371 | 36 | 8 | 5 | 2 220 |
| 7 | 0 | 1208 | 181 | 18 | 0 | 4 | 1 411 |
| 8 | 0 | 939 | 83 | 5 | 0 | 3 | 1 030 |
| 9 | 0 | 757 | 60 | 1 | 0 | 0 | 818 |
| 10 | 5 | 534 | 42 | 0 | 0 | 0 | 581 |
| 11 | 6 | 338 | 3 | 0 | 0 | 0 | 347 |
| 12 | 7 | 280 | 9 | 1 | 0 | 0 | 297 |
| 13 | 8 | 202 | 9 | 0 | 0 | 0 | 219 |
| 14 | 9 | 227 | 6 | 0 | 0 | 0 | 242 |
| 15 | 15 | 153 | 5 | 1 | 0 | 0 | 174 |
| 16 | 17 | 66 | 1 | 0 | 0 | 0 | 84 |
| 17 | 19 | 72 | 1 | 0 | 0 | 1 | 93 |
| 18 | 21 | 86 | 3 | 0 | 0 | 0 | 110 |
| 19 | 23 | 51 | 2 | 0 | 0 | 0 | 76 |
| 20 | 30 | 54 | 2 | 0 | 0 | 0 | 86 |
| >20 | 198 | 125 | 6 | 0 | 0 | 0 | 329 |
| 总数量 | 358 | 6 892 | 1 628 | 158 | 35 | 54 | 9 125 |
Table 4 Distribution of SSR repeat motif
重复基序数目 Number of repeated motifs | SSR位点数目 Number of SSR loci | 总数量 Total quantity | |||||
|---|---|---|---|---|---|---|---|
单核苷酸 Single nucleotide | 二核苷酸 Dinucleotide | 三核苷酸 Trinucleotide | 四核苷酸 Tetranucleotide | 五核苷酸 Pentanucleotide | 六核苷酸 Hexanucleotide | ||
| 5 | 0 | 0 | 844 | 96 | 27 | 41 | 1 008 |
| 6 | 0 | 1800 | 371 | 36 | 8 | 5 | 2 220 |
| 7 | 0 | 1208 | 181 | 18 | 0 | 4 | 1 411 |
| 8 | 0 | 939 | 83 | 5 | 0 | 3 | 1 030 |
| 9 | 0 | 757 | 60 | 1 | 0 | 0 | 818 |
| 10 | 5 | 534 | 42 | 0 | 0 | 0 | 581 |
| 11 | 6 | 338 | 3 | 0 | 0 | 0 | 347 |
| 12 | 7 | 280 | 9 | 1 | 0 | 0 | 297 |
| 13 | 8 | 202 | 9 | 0 | 0 | 0 | 219 |
| 14 | 9 | 227 | 6 | 0 | 0 | 0 | 242 |
| 15 | 15 | 153 | 5 | 1 | 0 | 0 | 174 |
| 16 | 17 | 66 | 1 | 0 | 0 | 0 | 84 |
| 17 | 19 | 72 | 1 | 0 | 0 | 1 | 93 |
| 18 | 21 | 86 | 3 | 0 | 0 | 0 | 110 |
| 19 | 23 | 51 | 2 | 0 | 0 | 0 | 76 |
| 20 | 30 | 54 | 2 | 0 | 0 | 0 | 86 |
| >20 | 198 | 125 | 6 | 0 | 0 | 0 | 329 |
| 总数量 | 358 | 6 892 | 1 628 | 158 | 35 | 54 | 9 125 |
| [1] | Yang JR, An Z, Li ZH, et al. Sesquiterpene coumarins from the roots of Ferula sinkiangensis and Ferula teterrima [J]. Chem Pharm Bull, 2006, 54(11): 1595-1598. |
| [2] | Xing YC, Li N, Zhou D, et al. Sesquiterpene coumarins from Ferula sinkiangensis act as neuroinflammation inhibitors [J]. Planta Med, 2017, 83(1/02): 135-142. |
| [3] | Li GZ, Li XJ, Cao L, et al. Steroidal esters from Ferula sinkiangensis [J]. Fitoterapia, 2014, 97: 247-252. |
| [4] | Ye BG, Wang S, Zhang L. Studies on the detoxification effects and acute toxicity of a mixture of Cis-sec-butyl-1-propoenyl disulphide and trans-sec-butyl-1-propoenyl disulphide isolated from crude essential oil of Ferula sinkiangensis K.M. Shen, a Chinese traditional herbal medicine [J]. Nat Prod Res, 2011, 25(12): 1161-1170. |
| [5] | Zhou YT, Xin F, Zhang GQ, et al. Recent advances on bioactive constituents in Ferula [J]. Drug Dev Res, 2017, 78(7): 321-331. |
| [6] | Bao XH, Li YP, Li Q, et al. Racemic norneolignans from the resin of Ferula sinkiangensis and their COX-2 inhibitory activity [J]. Fitoterapia, 2023, 164: 105341. |
| [7] | 李洁, 徐海燕, 贾晓光, 等. 阜康阿魏的化学成分及其生物活性研究进展 [J]. 新疆医科大学学报, 2012, 35(9): 1159-1161. |
| Li J, Xu HY, Jia XG, et al. Research progress on chemical constituents and biological activities of Ferula Fukang [J]. J Xinjiang Med Univ, 2012, 35(9): 1159-1161. | |
| [8] | Wang JC, Zheng Q, Wang HX, et al. Sesquiterpenes and sesquiterpene derivatives from Ferula: their chemical structures, biosynthetic pathways, and biological properties [J]. Antioxidants, 2024, 13(1): 7. |
| [9] | Khayat MT, Alharbi M, Ghazawi KF, et al. Ferula sinkiangensis (Chou-AWei, Chinese Ferula): traditional uses, phytoconstituents, biosynthesis, and pharmacological activities [J]. Plants, 2023, 12(4): 902. |
| [10] | Macrì R, Bava I, Scarano F, et al. In vitro evaluation of ferutinin Rich-Ferula communis L., ssp. glauca, root extract on Doxorubicin-induced cardiotoxicity: antioxidant properties and cell cycle modulation [J]. Int J Mol Sci, 2023, 24(16): 12735. |
| [11] | Mi Y, Jiao K, Xu JK, et al. Kellerin from Ferula sinkiangensis exerts neuroprotective effects after focal cerebral ischemia in rats by inhibiting microglia-mediated inflammatory responses [J]. J Ethnopharmacol, 2021, 269: 113718. |
| [12] | Arjmand Z, Hamburger M, Dastan D. Isolation and purification of terpenoid compounds from Ferula haussknechtii and evaluation of their antibacterial effects [J]. Nat Prod Res, 2023, 37(10): 1617-1624. |
| [13] | Wang JL, Sang CY, Wang J, et al. Sesquiterpene coumarins from Ferula sinkiangensis and their anti-pancreatic cancer effects [J]. Phytochemistry, 2023, 214: 113824. |
| [14] | Li GZ, Wang JC, Li XJ, et al. An unusual sesquiterpene coumarin from the seeds of Ferula sinkiangensis [J]. J Asian Nat Prod Res, 2016, 18(9): 891-896. |
| [15] | 樊丛照. 基于多组学分析的新疆阿魏开花调控机制研究 [D]. 北京: 北京协和医学院, 2024. |
| Fan CZ. Mechanism of flowering regulation in Ferula sinkiangensis based on multi-omies analysis [D]. Beijing: Peking Union Medical College, 2024. | |
| [16] | Tang XH, Li TJ, Hao ZY, et al. Identification of key genes involved in sesquiterpene synthesis in Nardostachys jatamansi based on transcriptome and component analysis [J]. Genes, 2024, 15(12): 1539. |
| [17] | Wang JC, Wang HJ, Zhang M, et al. Sesquiterpene coumarins from Ferula sinkiangensis K.M.Shen and their cytotoxic activities [J]. Phytochemistry, 2020, 180: 112531. |
| [18] | Wang JL, Zhao YM, Qiang Y. Chemical constituents from Ferula sinkiangensis and their chemotaxonomic significance [J]. Biochem Syst Ecol, 2022, 105: 104519. |
| [19] | Ahmed S, Zhan CS, Yang YY, et al. The transcript profile of a traditional Chinese medicine, Atractylodes lancea, revealing its sesquiterpenoid biosynthesis of the major active components [J]. PLoS One, 2016, 11(3): e0151975. |
| [20] | Vranová E, Coman D, Gruissem W. Network analysis of the MVA and MEP pathways for isoprenoid synthesis [J]. Annu Rev Plant Biol, 2013, 64: 665-700. |
| [21] | 陈建, 赵德刚. 植物萜类生物合成相关酶类及其编码基因的研究进展 [J]. 分子植物育种, 2004, 2(6): 757-764. |
| Chen J, Zhao DG. Research advances on the enzymes and their coding gene involved in plant terpene biosynthesis [J]. Mol Plant Breed, 2004, 2(6): 757-764. | |
| [22] | 郭连安, 潘媛, 谭均, 等. 不同生长年限木香转录组分析及倍半萜合成基因挖掘 [J]. 西南农业学报, 2024, 37(9): 2042-2050. |
| Guo LA, Pan Y, Tan J, et al. Transcriptome analysis of Aucklandiae lappa from different growth years and mining genes related to sesquiterpenes synthesis [J]. Southwest China J Agric Sci, 2024, 37(9): 2042-2050. | |
| [23] | Kim BR, Kim SU, Chang YJ. Differential expression of three 1-deoxy-D-xylulose-5-phosphate synthase genes in rice [J]. Biotechnol Lett, 2005, 27(14): 997-1001. |
| [24] | 曾珊珊, 高婷, 谷梦雅, 等. 基于黄棪(Camellia sinensis)基因组的DXS基因家族鉴定及表达 [J]. 应用与环境生物学报, 2024, 30(4): 811-817. |
| Zeng SS, Gao T, Gu MY, et al. Genome-wide identification and expression of the DXS gene family in Huangdan (Camellia sinensis) [J]. Chin J Appl Environ Biol, 2024, 30(4): 811-817. | |
| [25] | Zhang XW, Guo JW, Cheng FY, et al. Cytochrome P450 enzymes in fungal natural product biosynthesis [J]. Nat Prod Rep, 2021, 38(6): 1072-1099. |
| [26] | 高世玺, 戎梅, 彭俊祥, 等. 细胞色素P450在植物倍半萜类化合物生物合成中的作用研究进展 [J]. 药学学报, 2024, 59(2): 313-321. |
| Gao SX, Rong M, Peng JX, et al. Research progress on the role of cytochrome P450 in plant sesquiterpene biosynthesis [J]. Acta Pharm Sin, 2024, 59(2): 313-321. | |
| [27] | Chen DH, Liu CJ, Ye HC, et al. Ri-mediated transformation of Artemisia annua with a recombinant farnesyl diphosphate synthase gene for artemisinin production [J]. Plant Cell Tissue Organ Cult, 1999, 57(3): 157-162. |
| [28] | Ichinose H, Kitaoka T. Insight into metabolic diversity of the brown-rot basidiomycete Postia placenta responsible for sesquiterpene biosynthesis: semi-comprehensive screening of cytochrome P450 monooxygenase involved in protoilludene metabolism [J]. Microb Biotechnol, 2018, 11(5): 952-965. |
| [29] | Shoji T, Yuan L. ERF gene clusters: working together to regulate metabolism [J]. Trends Plant Sci, 2021, 26(1): 23-32. |
| [30] | Liu YH, Khan AR, Gan YB. C2H2 zinc finger proteins response to abiotic stress in plants [J]. Int J Mol Sci, 2022, 23(5): 2730. |
| [31] | 李超颖. 新疆阿魏化学成分及高效液相指纹图谱研究 [D]. 乌鲁木齐: 新疆大学, 2014. |
| Li CY. Studies of the chemical constituents and the HPLC fingerprints of Ferula sinkiangensis K.M.Shen [D]. Urumqi: Xinjiang University, 2014. | |
| [32] | 田再民, 张立平, 孙庆林, 等. 小麦数量性状基因定位研究进展 [J]. 内蒙古农业科技, 2007, 35(3): 68-71. |
| Tian ZM, Zhang LP, Sun QL, et al. Research review on wheat QTLs [J]. Inn Mong Agric Sci Technol, 2007, 35(3): 68-71. | |
| [33] | Fan CJ, Liu QY, Zeng BS, et al. Development of simple sequence repeat (SSR) markers and genetic diversity analysis in blackwood (Acacia melanoxylon) clones in China [J]. Silvae Genet, 2016, 65(1): 49-54. |
| [34] | Cregan PB, Akkaya MS, Bhagwat AA, et al. Length polymorphisms of simple sequence repeat (SSR) DNA as molecular markers in plants [M]// United Kingdom: CRC Press, Plant genome analysis, 1994: 47-56. |
| [35] | Kaur S, Pembleton LW, Cogan NO, et al. Transcriptome sequencing of field pea and faba bean for discovery and validation of SSR genetic markers [J]. BMC Genom, 2012, 13(1): 104. |
| [36] | Yang QW, Jiang YJ, Wang YP, et al. SSR loci analysis in transcriptome and molecular marker development in Polygonatum sibiricum . [J]. BioMed Res Int, 2022, 2022(1): 4237913. |
| [37] | Liu YL, Zhang PF, Song ML, et al. Transcriptome analysis and development of SSR molecular markers in Glycyrrhiza uralensis fisch [J]. PLoS One, 2015, 10(11): e0143017. |
| [1] | LIU Jian-guo, LIU Ge-er, GUO Ying-xin, WANG Bin, WANG Yu-kun, LU Jin-feng, HUANG Wen-ting, ZHU Yun-na. Integrate Transcriptomic and Metabolomic Analysis of Fruits Quality Differences between ‘Guiyou No. 1’ and ‘Shatianyou’ Pomelo (Citrus maxima) [J]. Biotechnology Bulletin, 2025, 41(9): 168-181. |
| [2] | LIU Ze-zhou, DUAN Nai-bin, YUE Li-xin, WANG Qing-hua, YAO Xing-hao, GAO Li-min, KONG Su-ping. Analysis of Wax Components and Screening of Wax-deficient Gene Ggl-1 in Garlic (Allium sativum L.) [J]. Biotechnology Bulletin, 2025, 41(9): 219-231. |
| [3] | YAN Meng-yang, LIANG Xiao-yang, DAI Jun-ang, ZHANG Yan, GUAN Tuan, ZHANG Hui, LIU Liang-bo, SUN Zhi-hua. Screening of Amoxicillin-degrading Bacteria and Study on Its Degradation Mechanisms [J]. Biotechnology Bulletin, 2025, 41(9): 314-325. |
| [4] | LU Yao, YUAN Ping-ping, JIN Xin, MAO Xiang-hong, FAN Xiang-bin, BAI Xiao-dong. Genetic Diversity Analysis and Fingerprinting of Wild Potato and Landraces Based on SSR Markers [J]. Biotechnology Bulletin, 2025, 41(9): 94-104. |
| [5] | PEI Hong-xia, WANG Lu-yao, LI Sheng-mei, GAO Jing-xia. Genetic Diversity of 220 Pepper Germplasm Resources Using SCoT, SRAP, and SSR Molecular Markers [J]. Biotechnology Bulletin, 2025, 41(8): 165-174. |
| [6] | BAI Yu-guo, LI Wan-di, LIANG Jian-ping, SHI Zhi-yong, LU Geng-long, LIU Hong-jun, NIU Jing-ping. Growth-promoting Mechanism of Trichoderma harzianum T9131 on Astragalus membranaceus Seedlings [J]. Biotechnology Bulletin, 2025, 41(8): 175-185. |
| [7] | WANG Yue-chen, HAN Xin-qi, WEI Wen-min, CUI Zhao-lan, LUO Yang-mei, CHEN Peng-ru, WANG Hai-gang, LIU long-long, ZHANG Li, WANG Lun. Biological Basis Study for Grain Shattering in Proso Millet and Identification of Genes Regulating Grain Shattering [J]. Biotechnology Bulletin, 2025, 41(7): 164-171. |
| [8] | ZHANG Yue, BI Yu, MU Xue-nan, ZHENG Zi-wei, WANG Zhi-gang, XU Wei-hui. Biocontrol Characteristics of Strain JB7 against Fusarium graminearum [J]. Biotechnology Bulletin, 2025, 41(7): 261-271. |
| [9] | DUAN Min-jie, LI Yi-fei, WANG Chun-ping, HUANG Ren-zhong, HUANG Qi-zhong, ZHANG Shi-cai. Association Analysis and Fingerprint Map Construction of Fruit Color Traits in Pepper via SSR Markers [J]. Biotechnology Bulletin, 2025, 41(7): 81-94. |
| [10] | LI Cheng-hua, DOU Fei-fei, REN Yu-zhao, LIU Cai-xia, LIU Feng-lou, WANG Zhang-jun, LI Qing-feng. Effect of Exogenous Salicylic Acid on Wheat Infested with Blumeria graminis f. sp. tritici and Its Transcriptome Analysis [J]. Biotechnology Bulletin, 2025, 41(7): 272-280. |
| [11] | GUO Xiu-juan, FENG Yu, WU Rui-xiang, WANG Li-qin, YANG Jian-chun. Transcriptome Analysis of the Effect of Ca 2+ Treatment on the Seed Germination of Flax [J]. Biotechnology Bulletin, 2025, 41(7): 139-149. |
| [12] | DUAN Yong-hong, YANG Xin, YU Guan-qun, XIA Jun-jun, SONG Lu-shuai, BAI Xiao-dong, PENG Suo-tang. Genetic Diversity and Principal Component Analysis of 125 Potato Germplasm Resources [J]. Biotechnology Bulletin, 2025, 41(6): 130-143. |
| [13] | LIU Yuan, ZHAO Ran, LU Zhen-fang, LI Rui-li. Research Progress in the Biological Metabolic Pathway and Functions of Plant Carotenoids [J]. Biotechnology Bulletin, 2025, 41(5): 23-31. |
| [14] | HU Ruo-qun, ZENG Jing-jing, LIANG Wan-feng, CAO Jia-yu, HUANG Xiao-wei, LIANG Xiao-ying, QIU Ming-yue, CHEN Ying. Integrated Transcriptome and Metabolome Analysis to Explore the Carotenoid Synthesis and Metabolism Mechanism in Anoectochilus roxburghii under Different Shading Conditions [J]. Biotechnology Bulletin, 2025, 41(5): 231-243. |
| [15] | TANG You, ZHAO Jun-wei, SUN Lan-xi, LI Xiang. Combined Application of Multi-omics Technologies in Plant Metabolic Pathway Resolution [J]. Biotechnology Bulletin, 2025, 41(4): 76-87. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||