








Biotechnology Bulletin ›› 2025, Vol. 41 ›› Issue (12): 95-105.doi: 10.13560/j.cnki.biotech.bull.1985.2025-0496
Previous Articles Next Articles
HU Wan-ke1,2( ), CHEN Yun-xia1,3, LUO Di-zhou4, WU Si-yu1, LI Jian-bo1, ZHAI Shao-lun5, JU Xiang-hong2, LIAO Ming6, WEI Wen-kang1(
), CHEN Yun-xia1,3, LUO Di-zhou4, WU Si-yu1, LI Jian-bo1, ZHAI Shao-lun5, JU Xiang-hong2, LIAO Ming6, WEI Wen-kang1( ), YU Jie-shi1(
), YU Jie-shi1( )
)
Received:2025-05-14
Online:2025-12-26
Published:2026-01-06
Contact:
WEI Wen-kang, YU Jie-shi
E-mail:2112204099@stu.gdou.edu.cn;weiwenkang@gdaas.cn;yujieshi@gdaas.cn
HU Wan-ke, CHEN Yun-xia, LUO Di-zhou, WU Si-yu, LI Jian-bo, ZHAI Shao-lun, JU Xiang-hong, LIAO Ming, WEI Wen-kang, YU Jie-shi. Establishment and Functional Validation of a Reverse Genetics System for the Chinese Influenza D Virus D/JY3002[J]. Biotechnology Bulletin, 2025, 41(12): 95-105.

Fig. 1 5′-and 3′-end sequences of genomic segments of influenza D virusesA-G illustrate 100 bp sequences at 5'- and 3'-end of genomic segments (PB2, PB1, P3, HEF, NP, M, and NS) of influenza D viruses that belong to different genetic lineages (D/OK, D/660, and D/Yama2019). The first and second rows in each sequence alignment diagram refer to terminal sequences of genomic segments from the D/OK and D/660 strains, respectively. The third row displays the terminal sequences of the D/JY3002 genomic segment, which have been reported but are incomplete. The fourth row presents the complete terminal sequence of the D/JY3002 genomic segment obtained in this study. The red box in the illustration highlights the sequences of the non-coding regions at both ends of genomic segments of influenza D viruses
| Number | Primer name | Primer sequence(5'-3') |
|---|---|---|
| 1 | D/JY3002-PB2-F | AGCATAAGCAGAGGATGTCACTACTATTAACGC |
| 2 | D/JY3002-PB2-R | CCGCCGGGTTATTAGCAGTAGCAAGAGGATTTTTTCAATGTG |
| 3 | D/JY3002-PB1-F | GGAGCATAAGCAGAGGATTTTATAACAATGGA |
| 4 | D/JY3002-PB1-R | CCGCCGGGTTATTAGCAGTAGCAAGAGGATTTTTC |
| 5 | D/JY3002-P3-F | AGCATAAGCAGGAGATTTAGAAATGTCTAGTAT |
| 6 | D/JY3002-P3-R | CCGCCGGGTTATTAGCAGTAGCAAGGAGATTTTTAA |
| 7 | D/JY3002-HEF-F | AGCATAAGCAGGAGATTTTCAAAGATGTTTTTG |
| 8 | D/JY3002-HEF-R | CCGCCGGGTTATTAGCAGTAGCAAGGAGATTTTTTCTAAGAT |
| 9 | D/JY3002-NP-F | AGCATAAGCAGGAGATTATTAAGCAATATGGAC |
| 10 | D/JY3002-NP-R | CCGCCGGGTTATTAGCAGTAGCAAGGAGATTTTTTGTTAAAT |
| 11 | D/JY3002-M-F | GGAGCATAAGCAGAGGATATTTTTGACGCAATG |
| 12 | D/JY3002-M-R | CCGCCGGGTTATTAGCAGTAGCAAGAGGATTTTTTCGCGA |
| 13 | D/JY3002-NS-F | GGAGCATAAGCAGGGGTGTACAATTTCAATATG |
| 14 | D/JY3002-NS-R | CCGCCGGGTTATTAGCAGTAGCAAGGGGTTTTTTCATACT |
| 15 | pHW2000-F | TACTGCTAATAACCCGGCGGCCCAAAATGCCG |
| 16 | pHW2000-PB2-R | TGACATCCTCTGCTTATGCTCCCCCCCAACTTCGGAGGTCGA |
| 17 | pHW2000-PB1-R | AAATCCTCTGCTTATGCTCCCCCCCAACTTCGGAGGTCGA |
| 18 | pHW2000-P3-R | CTAAATCTCCTGCTTATGCTCCCCCCCAACTTCGGAGGTCGA |
| 19 | pHW2000-HEF-R | GAAAATCTCCTGCTTATGCTCCCCCCCAACTTCGGAGGTCGA |
| 20 | pHW2000-NP-R | AATAATCTCCTGCTTATGCTCCCCCCCAACTTCGGAGGTCGA |
| 21 | pHW2000-M-R | ATATCCTCTGCTTATGCTCCCCCCCAAACTTCGGAGGTCGA |
| 22 | pHW2000-NS-R | TACACCCCTGCTTATGCTCCCCCCCAACTTCGGAGGTCGA |
Table 1 Primers for amplifying target fragments and linearization of vectors
| Number | Primer name | Primer sequence(5'-3') |
|---|---|---|
| 1 | D/JY3002-PB2-F | AGCATAAGCAGAGGATGTCACTACTATTAACGC |
| 2 | D/JY3002-PB2-R | CCGCCGGGTTATTAGCAGTAGCAAGAGGATTTTTTCAATGTG |
| 3 | D/JY3002-PB1-F | GGAGCATAAGCAGAGGATTTTATAACAATGGA |
| 4 | D/JY3002-PB1-R | CCGCCGGGTTATTAGCAGTAGCAAGAGGATTTTTC |
| 5 | D/JY3002-P3-F | AGCATAAGCAGGAGATTTAGAAATGTCTAGTAT |
| 6 | D/JY3002-P3-R | CCGCCGGGTTATTAGCAGTAGCAAGGAGATTTTTAA |
| 7 | D/JY3002-HEF-F | AGCATAAGCAGGAGATTTTCAAAGATGTTTTTG |
| 8 | D/JY3002-HEF-R | CCGCCGGGTTATTAGCAGTAGCAAGGAGATTTTTTCTAAGAT |
| 9 | D/JY3002-NP-F | AGCATAAGCAGGAGATTATTAAGCAATATGGAC |
| 10 | D/JY3002-NP-R | CCGCCGGGTTATTAGCAGTAGCAAGGAGATTTTTTGTTAAAT |
| 11 | D/JY3002-M-F | GGAGCATAAGCAGAGGATATTTTTGACGCAATG |
| 12 | D/JY3002-M-R | CCGCCGGGTTATTAGCAGTAGCAAGAGGATTTTTTCGCGA |
| 13 | D/JY3002-NS-F | GGAGCATAAGCAGGGGTGTACAATTTCAATATG |
| 14 | D/JY3002-NS-R | CCGCCGGGTTATTAGCAGTAGCAAGGGGTTTTTTCATACT |
| 15 | pHW2000-F | TACTGCTAATAACCCGGCGGCCCAAAATGCCG |
| 16 | pHW2000-PB2-R | TGACATCCTCTGCTTATGCTCCCCCCCAACTTCGGAGGTCGA |
| 17 | pHW2000-PB1-R | AAATCCTCTGCTTATGCTCCCCCCCAACTTCGGAGGTCGA |
| 18 | pHW2000-P3-R | CTAAATCTCCTGCTTATGCTCCCCCCCAACTTCGGAGGTCGA |
| 19 | pHW2000-HEF-R | GAAAATCTCCTGCTTATGCTCCCCCCCAACTTCGGAGGTCGA |
| 20 | pHW2000-NP-R | AATAATCTCCTGCTTATGCTCCCCCCCAACTTCGGAGGTCGA |
| 21 | pHW2000-M-R | ATATCCTCTGCTTATGCTCCCCCCCAAACTTCGGAGGTCGA |
| 22 | pHW2000-NS-R | TACACCCCTGCTTATGCTCCCCCCCAACTTCGGAGGTCGA |
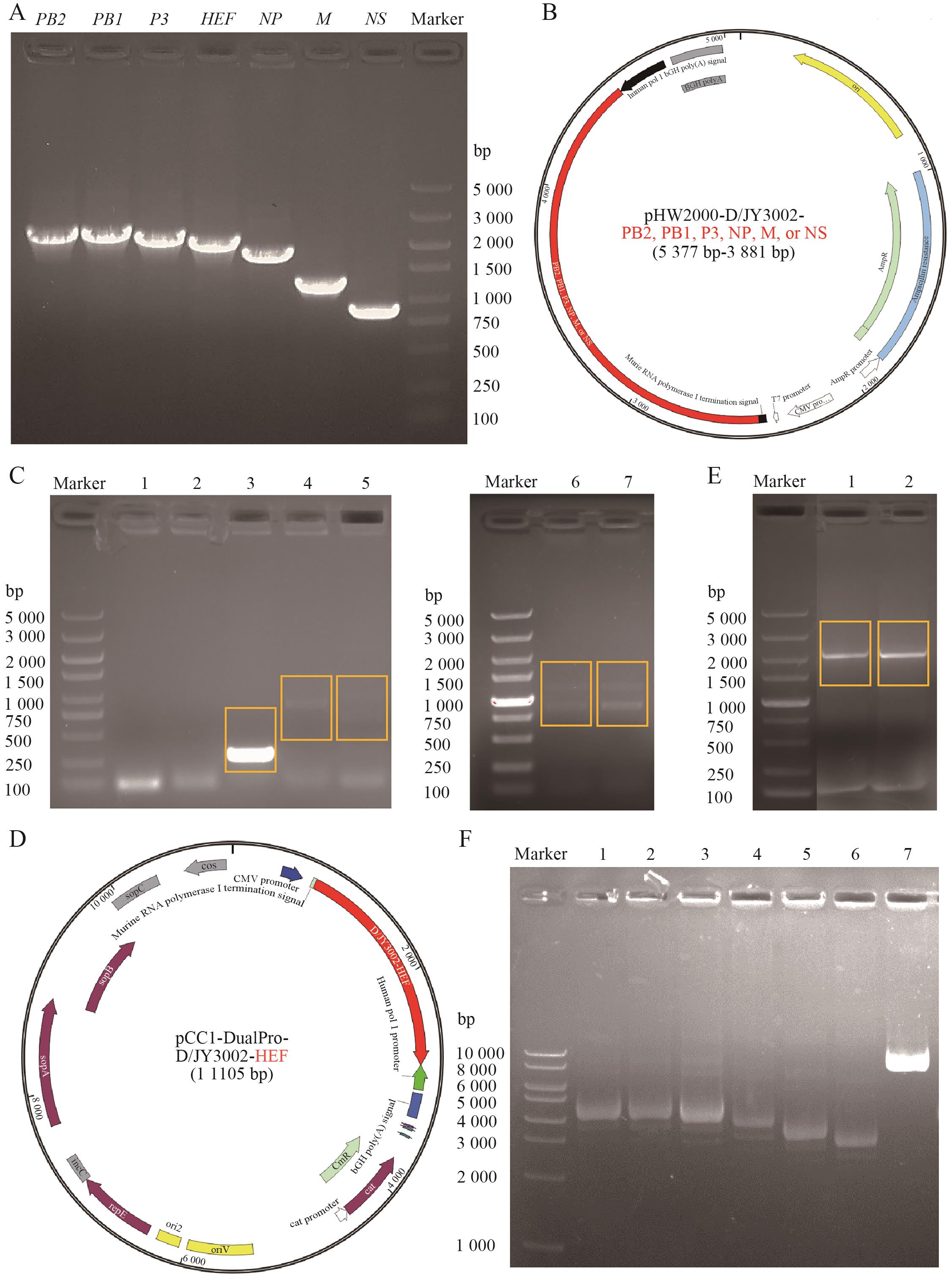
Fig. 2 Construction of bidirectional expression plasmids containing DNA fragments corresponding to the D/JY3002 genomic segmentsA: Gel electrophoresis plots of amplified DNA fragments corresponding to the full-length genomic segments PB2 (2 364 bp), PB1 (2 330 bp), P3 (2 195 bp), HEF (2 049 bp), NP (1 775 bp), M (1 219 bp) and NS (868 bp) of the D/JY3002. B: The map of bidirectional plasmid pHW2000 containing the DNA fragment corresponding to the full-length genomic segment PB2, PB1, P3, NP, M or NS of the D/JY3002.C: The DNA fragment corresponding to the D/JY3002 HEF genomic segment was ligated to the pHW2000 vector through seamless cloning technology. The colonies obtained after transformation were identified by PCR and analyzed by gel electrophoresis. Among them, lane 1 to 7 indicate 7 different colony samples respectively. D: The map of designed bidirectional plasmid pCC1-DualPro containing the DNA fragment corresponding to the full-length genomic segment HEF of the D/JY3002. E: The DNA fragment corresponding to the D/JY3002 HEF genomic segment was ligated to the pCC1-DualPro vector through seamless cloning technology. The colonies obtained after transformation were identified by PCR and analyzed by gel electrophoresis. Among them, lane 1 and 2 indicate 2 different colony samples. F: Gel electrophoresis analysis was performed on all the extracted bidirectional expression plasmids. Lane 1 to 7 are pHW2000-D/JY3002-PB2 (5 377 bp), -PB1 (5 343 bp), -P3 (5 208 bp), -NP (4 788 bp), -M (4 232 bp), -NS (3 881 bp), and pCC1-DualPro-D/JY3002-HEF (11 105 bp), respectively. Marker, used to indicate the size of the DNA fragment
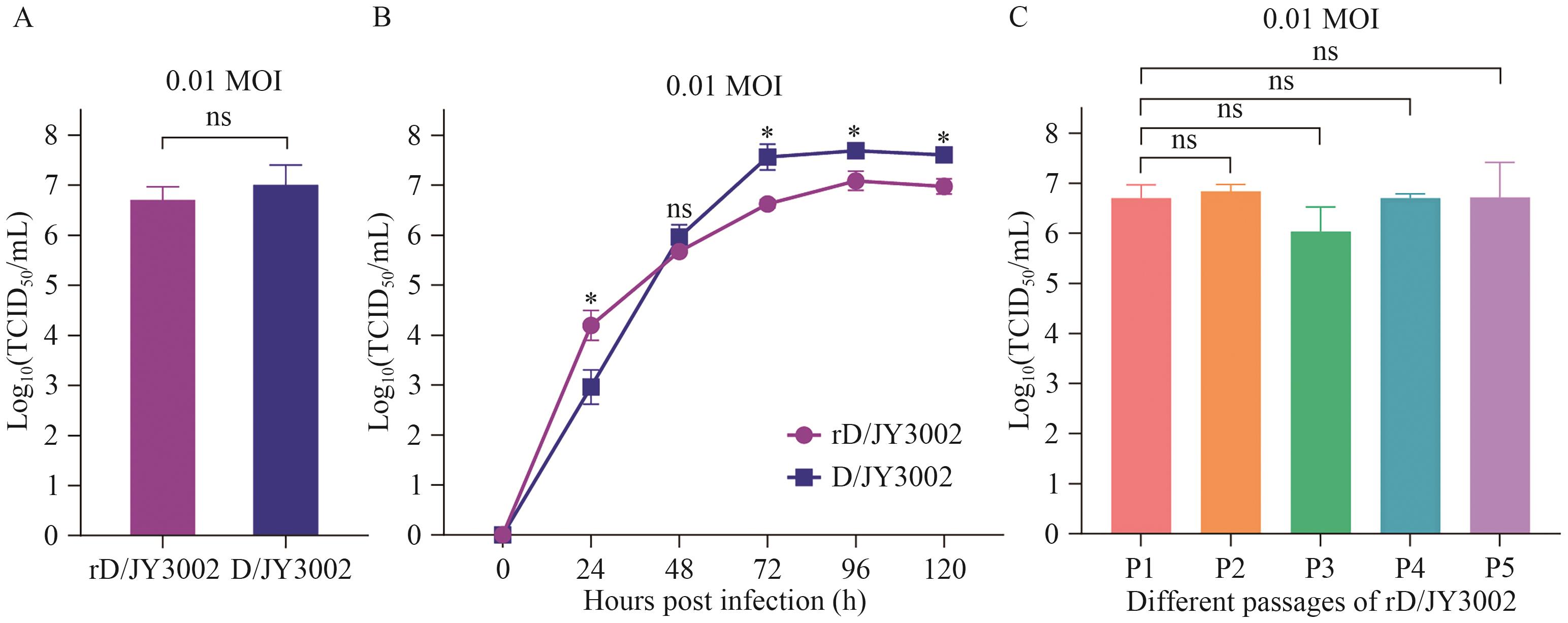
Fig. 3 Comparative analysis of the replication capacity and growth kinetics of artificially rescued rD/JY3002 and wild-type D/JY3002 influenza D virusesA: Replication titers (TCID50/mL) of the rescued rD/JY3002 and naturally isolated D/JY3002. B: Growth curves of rD/JY3002 and D/JY3002. C: Replication titers of different passages of rD/JY3002 (P1, P2, P3, P4 and P5). The infection dose was 0.01 MOI, and "ns" indicates no significant difference. * : P<0.05; ** : P<0.01 *** : P<0.001
Fig. 4 Artificial rescue of the rD/JY3002-GFP and expression analysis of the GFPA: Design and construction of the unidirectional expression plasmid pPolI-D/JY3002-PB1-240-GFP-240 (schematic diagram). B: Replication titers (TCID50/mL) of different passages of rD/JY3002-GFP viruses. C: Fluorescence images of MDCK cells infected with different passages of rD/JY3002-GFP viruses. Mock/Infected: Uninfected/Infected cells, and the infection dose is 0.01 MOI
Fig. 5 Construction of recombinant influenza D viruses containing the heterologous HEF gene and analysis of their antigenic cross-reactivityA: Schematic diagrams of recombinant influenza D viruses rD/JY3002, rD/JY3002-D/OK-HEF and rD/JY3002-D/660-HEF. B: Growth kinetics of rD/JY3002, rD/JY3002-D/OK-HEF and rD/JY3002-D/660-HEF on MDCK cells. C: HI titers of rabbit anti-D/JY3002 sera against recombinant influenza D viruses rD/JY3002, rD/JY3002-D/OK-HEF and rD/JY3002-D/660-HEF
| [1] | Hause BM, Ducatez M, Collin EA, et al. Isolation of a novel swine influenza virus from Oklahoma in 2011 which is distantly related to human influenza C viruses [J]. PLoS Pathog, 2013, 9(2): e1003176. |
| [2] | Hause BM, Collin EA, Liu RX, et al. Characterization of a novel influenza virus in cattle and swine: proposal for a new genus in the Orthomyxoviridae Family [J]. mBio, 2014, 5(2): e00031-14. |
| [3] | Collin EA, Sheng ZZ, Lang YK, et al. Cocirculation of two distinct genetic and antigenic lineages of proposed influenza D virus in cattle [J]. J Virol, 2015, 89(2): 1036-1042. |
| [4] | Ferguson L, Olivier AK, Genova S, et al. Pathogenesis of influenza D virus in cattle [J]. J Virol, 2016, 90(12): 5636-5642. |
| [5] | Yu JS, Li F, Wang D. The first decade of research advances in influenza D virus [J]. J Gen Virol, 2021, 102(1). |
| [6] | Trombetta CM, Marchi S, Marotta MG, et al. Detection of influenza D antibodies in dogs, Apulia Region, Italy, 2016 and 2023 [J]. Emerg Infect Dis, 2024, 30(5): 1045-1047. |
| [7] | Guan MH, Jacobson O, Sarafianos G, et al. Exposure of white-tailed deer in North America to influenza D virus [J]. Virology, 2022, 573: 111-117. |
| [8] | Yu JS, Hika B, Liu RX, et al. The hemagglutinin-esterase fusion glycoprotein is a primary determinant of the exceptional thermal and acid stability of influenza D virus [J]. mSphere, 2017, 2(4) |
| [9] | Huang C, Yu JS, Hause BM, et al. Emergence of new phylogenetic lineage of Influenza D virus with broad antigenicity in California, United States [J]. Emerg Microbes Infect, 2021, 10(1): 739-742. |
| [10] | Yu JS, Li TY, Wen ZY, et al. Identification of D/Yama2019 lineage-like influenza D virus in Chinese cattle [J]. Front Vet Sci, 2022, 9: 939456. |
| [11] | Yu JS, Wen ZY, Hu WK, et al. Influenza D virus infection in China, 2022-2023 [J]. Emerg Microbes Infect, 2024, 13(1): 2343907. |
| [12] | Yu JS, Liu RX, Zhou B, et al. Development and characterization of a reverse-genetics system for influenza D virus [J]. J Virol, 2019, 93(21): e01186-19. |
| [13] | Ishida H, Murakami S, Kamiki H, et al. Establishment of a reverse genetics system for influenza D virus [J]. J Virol, 2020, 94(10): e01767-19. |
| [14] | Holwerda M, Laloli L, Wider M, et al. Establishment of a reverse genetic system from a bovine derived influenza D virus isolate [J]. Viruses, 2021, 13(3): 502. |
| [15] | Murakami S, Sato R, Ishida H, et al. Influenza D virus of new phylogenetic lineage, Japan [J]. Emerg Infect Dis, 2020, 26(1): 168-171. |
| [16] | Hoffmann E, Neumann G, Kawaoka Y, et al. A DNA transfection system for generation of influenza A virus from eight plasmids [J]. Proc Natl Acad Sci U S A, 2000, 97(11): 6108-6113. |
| [17] | Hoffmann E, Mahmood K, Yang CF, et al. Rescue of influenza B virus from eight plasmids [J]. Proc Natl Acad Sci U S A, 2002, 99(17): 11411-11416. |
| [18] | Crescenzo-Chaigne B, van der Werf S. Rescue of influenza C virus from recombinant DNA [J]. J Virol, 2007, 81(20): 11282-11289. |
| [19] | Pleschka S, Jaskunas R, Engelhardt OG, et al. A plasmid-based reverse genetics system for influenza A virus [J]. J Virol, 1996, 70(6): 4188-4192. |
| [20] | Fodor E, Devenish L, Engelhardt OG, et al. Rescue of influenza a virus from recombinant DNA [J]. J Virol, 1999, 73(11): 9679-9682. |
| [21] | Noda T. Selective genome packaging mechanisms of influenza a viruses [J]. Cold Spring Harb Perspect Med, 2020: a038497. |
| [22] | Jiang WM, Wang SC, Peng C, et al. Identification of a potential novel type of influenza virus in Bovine in China [J]. Virus Genes, 2014, 49(3): 493-496. |
| [23] | Zhai SL, Zhang H, Chen SN, et al. Influenza D virus in animal species in Guangdong Province, Southern China [J]. Emerg Infect Dis, 2017, 23(8): 1392-1396. |
| [24] | He WT, Lu M, Xing G, et al. Emergence and adaptive evolution of influenza D virus [J]. Microb Pathog, 2021, 160: 105193. |
| [25] | 余界石, 魏文康. 丁型流感病毒在中国的流行状况及研究进展 [J]. 广东农业科学, 2022, 49(11): 1-8. |
| Yu JS, Wei WK. Epidemiology and research advances of influenza D virus in China [J]. Guangdong Agric Sci, 2022, 49(11): 1-8. | |
| [26] | Gao HB, Sun WY, Lu PY, et al. First isolation of influenza D virus from cattle in Northeast China [J]. Microbiol Spectr, 2024, 12(9): e00374-24. |
| [27] | Lim EH, Lim SI, Kim MJ, et al. First detection of influenza D virus infection in cattle and pigs in the republic of Korea [J]. Microorganisms, 2023, 11(7): 1751. |
| [28] | Nakatsu S, Murakami S, Shindo K, et al. Influenza C and D viruses package eight organized ribonucleoprotein complexes [J]. J Virol, 2018, 92(6): e02084-17. |
| [29] | Nakatsu S, Sagara H, Sakai-Tagawa Y, et al. Complete and incomplete genome packaging of influenza a and B viruses [J]. mBio, 2016, 7(5): e01248-16. |
| [1] | DOU Shuo, DING Ruo-xi, SUN Xing, GUO Wen-jing, KONG Wen-hui, YUAN Jing-xian, ZHANG Dong-mei, WANG Xing-fen, MA Zhi-ying, WU jin-hua, WU Li-zhu. Construction and Application Study of a General Vector pCamRUBY for Visual Screening of Transgenes [J]. Biotechnology Bulletin, 2025, 41(12): 66-73. |
| [2] | WANG Jing, CHANG Xue-rui, JIA Xu, HUANG Jia-xin, WANG Tian-tian, LIANG Yan-ping. Cloning and Fuctional Analysis of CaUBC38 Gene in Pepper [J]. Biotechnology Bulletin, 2025, 41(10): 242-252. |
| [3] | ZHANG Jing-an, HU Xiao-long, CAO Bei-bei, LIAO Min, SHU Chang-long, ZHANG Jie, WANG Kui, CAO Hai-qun. Construction and Characterization of Rapid Visual Expression Vector for Bacillus thuringiensis [J]. Biotechnology Bulletin, 2025, 41(1): 95-102. |
| [4] | WANG Yu-shu, ZHAO Lin-lin, ZHAO Shuang, HU Qi, BAI Hui-xia, WANG Huan, CAO Ye-ping, FAN Zhen-yu. Cloning and Expression Analysis of BrCYP83B1 Gene in Chinese Cabbage [J]. Biotechnology Bulletin, 2024, 40(6): 152-160. |
| [5] | ZHANG Zhen, LI Qing, XU Jing, CHEN Kai-yuan, ZHANG Chun-zhi, ZHU Guang-tao. Construction and Application of Potato Mitochondrial Targeted Expression Vector [J]. Biotechnology Bulletin, 2024, 40(5): 66-73. |
| [6] | MEI Xian-jun, SONG Hui-yang, LI Jing-hao, MEI Chao, SONG Qian-na, FENG Rui-yun, CHEN Xi-ming. Cloning and Expression Analysis of StDof5 Gene in Potato [J]. Biotechnology Bulletin, 2024, 40(3): 181-192. |
| [7] | ZHOU Jia-wei, WU Zhi-qiang. Construction Method of mitoTALENs Mitochondrial Gene Editing Vector in Plants [J]. Biotechnology Bulletin, 2024, 40(10): 172-180. |
| [8] | LIU Meng-meng, HAN Li-jun, LIU Bao-ling, XUE Jin-ai, LI Run-zhi. Cloning and Expression Analysis of GhSDP1 and Its Promoter in Gossypium hirsutum [J]. Biotechnology Bulletin, 2022, 38(2): 34-43. |
| [9] | WU Kun-kun, XU Xing, JI Ce, REN Jian-feng, LI Wei-ming, ZHANG Qing-hua. Eukaryotic Expression Vector Construction of Danio rerio notch3 Gene and Its Expression Analysis [J]. Biotechnology Bulletin, 2022, 38(1): 179-186. |
| [10] | HU Zi-yao, DAI Pei-hong, LIU Chao, Madina Mulati, WANG Qian, Wugalihan Abuduwili, ZHAO Yi, SUN Ling, XU Shi-jia, LI Yue. Molecular Cloning,Expression and VIGS Construction of a Small GTP-binding Protein Gene GhROP3 in Gossypium hirsutum [J]. Biotechnology Bulletin, 2021, 37(9): 106-113. |
| [11] | GAO Peng-fei, XI Fei-hu, ZHANG Ze-yu, HU Kai-qiang, CHEN Kai, WEI Wen-tao, DING Jia-zhi, GU Lian-feng. Research Progress of Plant VIGS Technology and Its Application in Forestry Science [J]. Biotechnology Bulletin, 2021, 37(5): 141-153. |
| [12] | GONG Yuan-yong, ZHAO Li-hua, YAN Fei, GUO Shu-qiao, SHU Hong-mei, NI Wan-chao. Construction and Transformation of Borago officinalis BoD6D Gene Vector [J]. Biotechnology Bulletin, 2021, 37(3): 227-232. |
| [13] | SUN Jing-shuang, HU Rui-yang, ZHENG Guang-shun, MA Wen-jun, XU Yan, WANG Jun-hui. Research Progress and Prospect of Plant Genetic Transformation Mediated by Nano-gene Vector [J]. Biotechnology Bulletin, 2021, 37(2): 162-173. |
| [14] | HU Xiao, WANG Bao-bao, DOU Shao-hua, JIANG Nan, FU Chang-zhen, JIN Hang, GAO Feng-shan. Construction of a Eukaryotic Expression Vector of SLA-2 Gene from Yantai Black Pigs and Its Expression [J]. Biotechnology Bulletin, 2021, 37(10): 143-151. |
| [15] | WANG Cai-xia, DU Fang-yuan, LIN Xiang-mei, Grzegorz Wozniakowski, WANG Qin, FENG Chun-yan, WU Shao-qiang. Generation of a Vero Cell Line Stably Expressing African Swine Fever Virus P54 Protein [J]. Biotechnology Bulletin, 2020, 36(5): 139-144. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||