








Biotechnology Bulletin ›› 2025, Vol. 41 ›› Issue (12): 66-73.doi: 10.13560/j.cnki.biotech.bull.1985.2024-1133
Previous Articles Next Articles
DOU Shuo1( ), DING Ruo-xi1, SUN Xing2, GUO Wen-jing1, KONG Wen-hui1, YUAN Jing-xian1, ZHANG Dong-mei1, WANG Xing-fen1, MA Zhi-ying1, WU jin-hua1(
), DING Ruo-xi1, SUN Xing2, GUO Wen-jing1, KONG Wen-hui1, YUAN Jing-xian1, ZHANG Dong-mei1, WANG Xing-fen1, MA Zhi-ying1, WU jin-hua1( ), WU Li-zhu1(
), WU Li-zhu1( )
)
Received:2024-11-24
Online:2025-12-26
Published:2026-01-06
Contact:
WU jin-hua, WU Li-zhu
E-mail:1366761040@qq.com;nxywjh@126.com;wulizhu2008@163.com
DOU Shuo, DING Ruo-xi, SUN Xing, GUO Wen-jing, KONG Wen-hui, YUAN Jing-xian, ZHANG Dong-mei, WANG Xing-fen, MA Zhi-ying, WU jin-hua, WU Li-zhu. Construction and Application Study of a General Vector pCamRUBY for Visual Screening of Transgenes[J]. Biotechnology Bulletin, 2025, 41(12): 66-73.
引物名称 Primer name | 引物序列 Primer sequence (5′-3′) |
|---|---|
| RUBYF | CCGTGCACATCAACGACACC |
| RUBYR | GGAGGTGAACTTGTAGGAGC |
| AtActinF | CTGGTGATGGTGTGTCTCACAC |
| AtActinR | GCGATCCAGACACTGTACTTCC |
| GhActinF | GATCCTTCCTGATATCCACATCG |
| GhActinR | GGCACACTGGTGTTATGGTTGGG |
Table 1 Primers' names and sequence information
引物名称 Primer name | 引物序列 Primer sequence (5′-3′) |
|---|---|
| RUBYF | CCGTGCACATCAACGACACC |
| RUBYR | GGAGGTGAACTTGTAGGAGC |
| AtActinF | CTGGTGATGGTGTGTCTCACAC |
| AtActinR | GCGATCCAGACACTGTACTTCC |
| GhActinF | GATCCTTCCTGATATCCACATCG |
| GhActinR | GGCACACTGGTGTTATGGTTGGG |
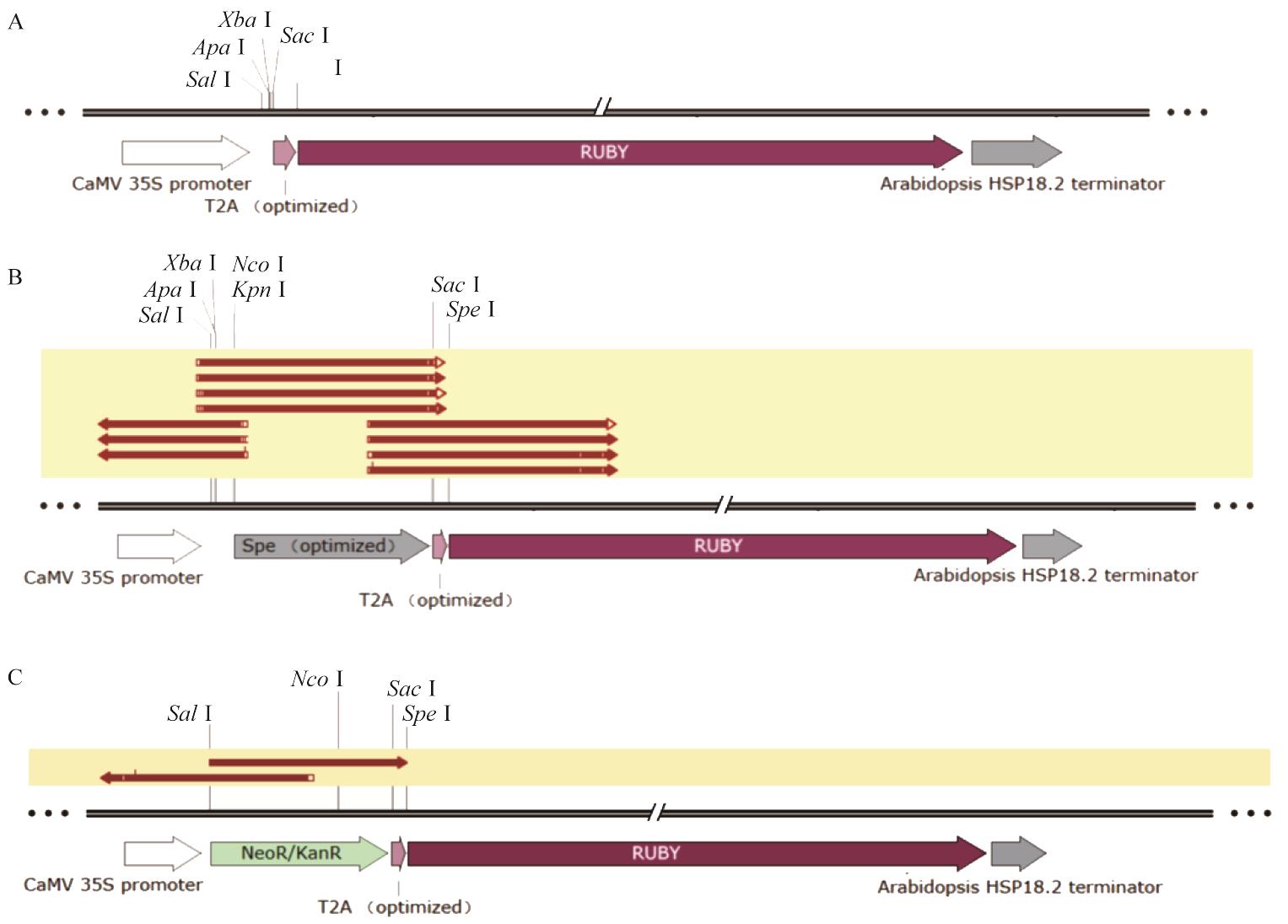
Fig. 1 Schematic diagram of vector results and sequencing alignment resultsA: pCamRUBY vector profile. B: pCamSpeRUBY vector profile and sequencing alignment results. C: pCamKanRUBY vector profile and sequencing alignment results

Fig. 2 Transgenic Arabidopsisthaliana screeningA: Red Arabidopsis were obtained through Spe resistance screening. B: Red Arabidopsis were directly screened with naked eye on nutrient soil. C: T2 generation pCamSpeRUBY transgenic Arabidopsis. The arrow indicates the transgenic positive plants. Bar=1 cm

Fig. 3 Phenotype of transgenic A. thalianaA: Phenotype of wild type and transgenic Arabidopsis inflorescence. B: Phenotype of bolting wild type and transgenic Arabidopsis. C: Phenotype of wild type and transgenic Arabidopsis plants. D: Phenotype of wild type and transgenic Arabidopsis seeds. E: PCR identification of RUBY gene in transgenic Arabidopsis. F: PCR identification of AtActin gene in in transgenic Arabidopsis. M: DNA maker DL2000. 1-2: Transgenic Arabidopsis plants obtained through resistance screening. 3-5: Transgenic Arabidopsis plants obtained by direct seeding. P: Plasmid pCamRUBY. WT: Wild-type control Arabidopsis plant. c: Blank control. Bar=1 cm
荚果编号 No. of pod | 种子数 Number of seeds | 红色种子数 Number of red seeds | 黄褐色种子数 Number of tawny seeds |
|---|---|---|---|
| 1 | 28 | 21 | 7 |
| 2 | 22 | 17 | 5 |
| 3 | 23 | 18 | 5 |
| 4 | 32 | 25 | 7 |
| 5 | 20 | 15 | 5 |
| 6 | 19 | 15 | 4 |
| 7 | 25 | 18 | 7 |
| 8 | 29 | 21 | 8 |
| 9 | 24 | 18 | 6 |
| 10 | 26 | 19 | 7 |
| Total | 248 | 187 | 61 |
Table 2 Separation ratio statistics of transgenic Arabidopsis seeds
荚果编号 No. of pod | 种子数 Number of seeds | 红色种子数 Number of red seeds | 黄褐色种子数 Number of tawny seeds |
|---|---|---|---|
| 1 | 28 | 21 | 7 |
| 2 | 22 | 17 | 5 |
| 3 | 23 | 18 | 5 |
| 4 | 32 | 25 | 7 |
| 5 | 20 | 15 | 5 |
| 6 | 19 | 15 | 4 |
| 7 | 25 | 18 | 7 |
| 8 | 29 | 21 | 8 |
| 9 | 24 | 18 | 6 |
| 10 | 26 | 19 | 7 |
| Total | 248 | 187 | 61 |
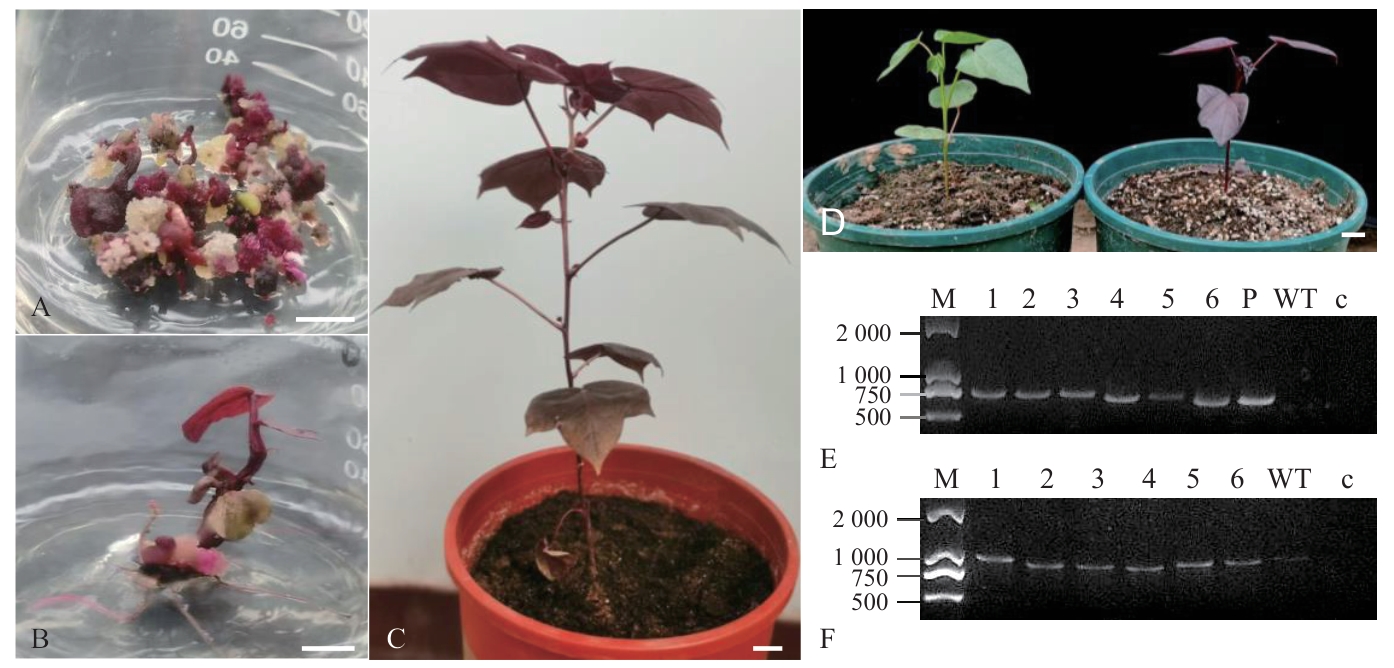
Fig. 4 Phenotypic observation of genetically transformed cotton calli and plants and PCR identifications of positive plantsA: Red transgenic cotton callus. B: Red transgenic cotton seedlings. C: Red transgenic cotton plants. D: Phenotype of wild type and T1 transgenic plants. E: PCR identification of RUBY gene in transgenic cotton. F: PCR identification of GhActin gene in transgenic cotton. M: DNA marker DL2000; 1-3: T0 transgenic cotton plants; 4-6: T1 transgenic cotton plants; P: plasmid pCamRUBY; WT: wild-type cotton J668; c: blank control; Bar=1 cm
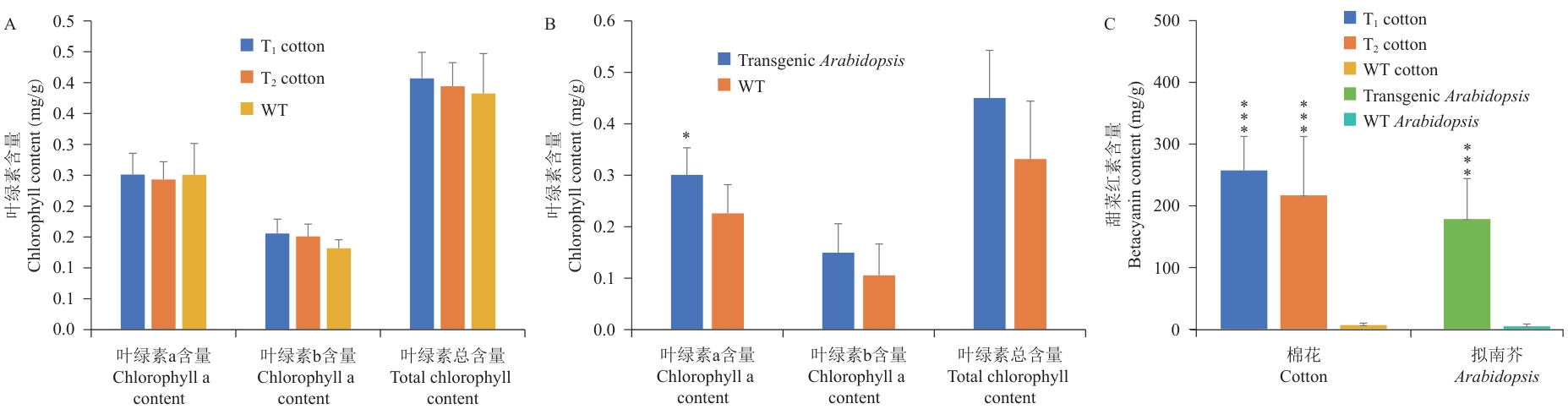
Fig. 5 Content detection of chlorophyll and betacyanin in transgenic cotton and ArabidopsisA: Determination of chlorophyll content in transgenic cotton. B: Determination of chlorophyll content in transgenic A. thaliana. C: Determination of betacyanin content in transgenic plants. *P<0.05; ***P<0.001
| [1] | 翟琼, 陈容钦, 梁晓华, 等. 一种花生快速遗传转化方法的建立与应用 [J]. 植物学报, 2022, 57(3): 327-339. |
| Zhai Q, Chen RQ, Liang XH, et al. Establishment and application of a rapid genetic transformation method for peanut [J]. Chin Bull Bot, 2022, 57(3): 327-339. | |
| [2] | Tang YP, Shen XY, Deng X, et al. Establishment of an efficient Agrobacterium-mediated transformation system for chilli pepper and its application in genome editing [J]. Plant Biotechnol J, 2025, 23: 4752-4754. |
| [3] | 柴燕文, 马晖玲, 谢小冬, 等. pBI121-Lyz-GFP表达载体的构建及其在和田苜蓿愈伤组织中的转化 [J]. 农业生物技术学报, 2008, 16(5): 842-846. |
| Chai YW, Ma HL, Xie XD, et al. Construction of pBI121-lyz-GFP expression vector and its transformation in callus of Medicago sativa [J]. J Agric Biotechnol, 2008, 16(5): 842-846. | |
| [4] | 张献龙, 李涛, 孙济中. 抗生素对棉花愈伤组织诱导和生长的影响 [J]. 华中农业大学学报, 1996, 15(2): 123-126. |
| Zhang XL, Li T, Sun JZ. The effect of antibiotics on the callus induction and growth in upland cotton (Gossypium hirsutum L.) [J]. J Huazhong Cent China Agric Univ, 1996, 15(2): 123-126. | |
| [5] | 刘巧泉, 陈秀花, 王兴稳, 等. 一种快速检测转基因水稻中潮霉素抗性的简易方法 [J]. 农业生物技术学报, 2001, 9(3): 264-268, 308. |
| Liu QQ, Chen XH, Wang XW, et al. A rapid simple method of assaying hygromycin resistance in transgenic rice plants [J]. J Agric Biotechnol, 2001, 9(3): 264-268, 308. | |
| [6] | Borovinskaya MA, Shoji S, Fredrick K, et al. Structural basis for hygromycin B inhibition of protein biosynthesis [J]. RNA, 2008, 14(8): 1590-1599. |
| [7] | 肖国樱, 唐俐, 袁定阳, 等. 转Bar基因抗除草剂两系杂交早稻恢复系Bar 68-1的培育研究 [J]. 杂交水稻, 2007, 22(6): 57-61. |
| Xiao GY, Tang L, Yuan DY, et al. Studies on development of bar 68-1, a bar-transgenic restorer line with herbicide resistance of two-line early hybrid rice [J]. Hybrid Rice, 2007, 22(6): 57-61. | |
| [8] | Subach FV, Patterson GH, Manley S, et al. Photoactivatable mCherry for high-resolution two-color fluorescence microscopy [J]. Nat Methods, 2009, 6(2): 153-159. |
| [9] | 陶曼芝, 吴席, 朱香豫, 等. 拟南芥ProWRKY12-GUS转基因植株的构建及其染色分析 [J]. 合肥工业大学学报: 自然科学版, 2022, 45(9): 1248-1251. |
| Tao MZ, Wu X, Zhu XY, et al. Construction and staining analysis of Arabidopsis thaliana ProWRKY12-GUS transgenic plants [J]. J Hefei Univ Technol Nat Sci, 2022, 45(9): 1248-1251. | |
| [10] | 崔志芳, 邹玉红, 季爱云. 绿色荧光蛋白研究的三个里程碑——2008年诺贝尔化学奖简介 [J]. 自然杂志, 2008, 30(6): 324-328. |
| Cui ZF, Zou YH, Ji AY. Three landmarks during the research of the green fluorescent protein(GFP): a brief introduction to the 2008 Nobel prize in chemistry [J]. Chin J Nat, 2008, 30(6): 324-328. | |
| [11] | Magness ST, Bataller R, Yang L, et al. A dual reporter gene transgenic mouse demonstrates heterogeneity in hepatic fibrogenic cell populations [J]. Hepatology, 2004, 40(5): 1151-1159. |
| [12] | Uenaka H, Kadota A. Functional analyses of the Physcomitrella patens phytochromes in regulating chloroplast avoidance movement [J]. Plant J, 2007, 51(6): 1050-1061. |
| [13] | 朱向玲, 吴旭铭, 王卉卉, 等. 基于Cre/Loxp系统的Csf1r-CreERT2R26REYFP报告基因小鼠的构建及效率检测 [J]. 安徽医科大学学报, 2024, 59(7): 1175-1180. |
| Zhu XL, Wu XM, Wang HH, et al. Construction and efficiency detection of Csf1r-CreERT2 R26REYFP reporter gene mouse based on Cre/Loxp system [J]. Acta Univ Med Anhui, 2024, 59(7): 1175-1180. | |
| [14] | He YB, Zhang T, Sun H, et al. A reporter for noninvasively monitoring gene expression and plant transformation [J]. Hortic Res, 2020, 7(1): 152. |
| [15] | 黄晨, 张威, 任红旭. 植物中甜菜色素的研究进展 [J]. 西北植物学报, 2023, 43(12): 2149-2160. |
| Huang C, Zhang W, Ren HX. Research progress of betalain in plants [J]. Acta Bot Boreali Occidentalia Sin, 2023, 43(12): 2149-2160. | |
| [16] | Liu J, Li H, Hong CH, et al. Quantitative RUBY reporter assay for gene regulation analysis [J]. Plant Cell Environ, 2024, 47(10): 3701-3711. |
| [17] | Šola K, Gilchrist EJ, Ropartz D, et al. RUBY, a putative galactose oxidase, influences pectin properties and promotes cell-to-cell adhesion in the seed coat epidermis of Arabidopsis [J]. Plant Cell, 2019, 31(4): 809-831. |
| [18] | Wang D, Zhong Y, Feng B, et al. The RUBY reporter enables efficient haploid identification in maize and tomato [J]. Plant Biotechnol J, 2023, 21(8): 1707-1715. |
| [19] | 林凯, 张育华, 柴霞, 等. 发根农杆菌介导的星油藤毛状根遗传转化体系建立 [J]. 植物生理学报, 2023, 59(2): 373-382. |
| Lin K, Zhang YH, Chai X, et al. Establishment of an Agrobacterium rhizogenes-mediated genetic transformation system of hairy roots in Sacha inchi (Plukenetia volubilis L.) [J]. Plant Physiol J, 2023, 59(2): 373-382. | |
| [20] | 吴立柱, 安叶芝, 张洁, 等. 基于核糖体跳跃的多顺反子共表达技术在拟南芥上的应用 [J]. 农业生物技术学报, 2019, 27(7): 1141-1148. |
| Wu LZ, An YZ, Zhang J, et al. Application of ribosomal skipping based polycistronic co-expression technique in Arabidopsis thaliana [J]. J Agric Biotechnol, 2019, 27(7): 1141-1148. | |
| [21] | Momose F, Morikawa Y. Polycistronic expression of the influenza a virus RNA-dependent RNA polymerase by using the Thosea asigna virus 2A-like self-processing sequence [J]. Front Microbiol, 2016, 7: 288. |
| [22] | 吴立柱, 王省芬, 李喜焕, 等. 通用型植物表达载体pCamE的构建及功能验证 [J]. 农业生物技术学报, 2014, 22(6): 661-671. |
| Wu LZ, Wang XF, Li XH, et al. Construction and function identification of universal plant expression vector pCamE [J]. J Agric Biotechnol, 2014, 22(6): 661-671. | |
| [23] | 马峙英, 吴立柱, 王省芬, 等. 植物表达载体pCamE及其在基因转化中的应用: CN102816790A [P]. 2012-12-12. |
| Ma ZY, Wu LZ, Wang XF, et al. Plant expression vector pCamE and application thereof to gene transformation: CN102816790A [P]. 2012-12-12. | |
| [24] | Ali I, Sher H, Ali A, et al. Simplified floral dip transformation method of Arabidopsis thaliana [J]. J Microbiol Meth, 2022, 197: 106492. |
| [25] | 鲍红帅, 尚红燕, 王国宁, 等. 不同陆地棉品种愈伤组织诱导比较及遗传转化验证 [J]. 河北农业大学学报, 2023, 46(1): 31-36. |
| Bao HS, Shang HY, Wang GN, et al. Comparison of callus induction ability of different upland cotton varieties and verification of genetic transformation [J]. J Hebei Agric Univ, 2023, 46(1): 31-36. | |
| [26] | 段秋霞, 李定金, 段振华, 等. 红心火龙果酒颜色稳定性影响因素的研究 [J]. 食品研究与开发, 2020, 41(24): 130-136. |
| Duan QX, Li DJ, Duan ZH, et al. Study on the factors affecting the color stability of red heart pitaya wine [J]. Food Res Dev, 2020, 41(24): 130-136. | |
| [27] | Liu HQ, Zhao JY, Chen FF, et al. Improving Agrobacterium tumefaciens-mediated genetic transformation for gene function studies and mutagenesis in cucumber (Cucumis sativus L.) [J]. Genes, 2023, 14(3): 601. |
| [28] | 葛杰, 何玉池, 展帆, 等. 多倍体水稻DHF-CBS基因的抗旱性鉴定 [J]. 湖北农业科学, 2024, 63(9): 216-220. |
| Ge J, He YC, Zhan F, et al. Identification of drought resistance of polyploid Oryza sativa L. DHF-CBS gene [J]. Hubei Agric Sci, 2024, 63(9): 216-220. | |
| [29] | Bai MY, Yuan JH, Kuang HQ, et al. Generation of a multiplex mutagenesis population via pooled CRISPR-Cas9 in soya bean [J]. Plant Biotechnol J, 2020, 18(3): 721-731. |
| [30] | Berthomieu P, Béclin C, Charlot F, et al. Routine transformation of rapid cycling cabbage (Brassica oleracea)—molecular evidence for regeneration of chimeras [J]. Plant Sci, 1994, 96(1/2): 223-235. |
| [31] | Mathews H, Dewey V, Wagoner W, et al. Molecular and cellular evidence of chimaeric tissues in primary transgenics and elimination of chimaerism through improved selection protocols [J]. Transgenic Res, 1998, 7(2): 123-129. |
| [32] | Faize M, Faize L, Burgos L. Using quantitative real-time PCR to detect chimeras in transgenic tobacco and apricot and to monitor their dissociation [J]. BMC Biotechnol, 2010, 10: 53. |
| [33] | Wu YH, Li J, Wang YL, et al. Development and application of a general plasmid reference material for GMO screening [J]. Plasmid, 2016, 87-88: 28-36. |
| [1] | HU Lu, WANG Kai, XU Jing-yi, YE Li-hui, WANG Yong-fei, WANG Li-hua, LI Jie-qin. Research Progress in Genetic Transformation Technologies of Maize and Sorghum [J]. Biotechnology Bulletin, 2025, 41(9): 32-43. |
| [2] | DENG Mei-bi, YAN Lang, ZHAN Zhi-tian, ZHU Min, HE Yu-bing. Efficient CRISPR Gene Editing in Rice Assisted by RUBY [J]. Biotechnology Bulletin, 2025, 41(8): 65-73. |
| [3] | PEI Jing-qi, ZHAO Meng-ran, HUANG Chen-yang, WU Xiang-li. Discovery and Verification of a Functional Gene Influencing the Growth and Development of Pleurotus ostreatus [J]. Biotechnology Bulletin, 2025, 41(6): 327-334. |
| [4] | YANG Chao-jie, ZHANG Lan, CHEN Hong, HUANG Juan, SHI Tao-xiong, ZHU Li-wei, CHEN Qing-fu, LI Hong-you, DENG Jiao. Functional Identification of the Transcription Factor Gene FtbHLH3 in Regulating Flavonoid Biosynthesis in Fagopyrum tataricum [J]. Biotechnology Bulletin, 2025, 41(4): 134-144. |
| [5] | WEN Bo-lin, WAN Min, HU Jian-jun, WANG Ke-xiu, JING Sheng-lin, WANG Xin-yue, ZHU Bo, TANG Ming-xia, LI Bing, HE Wei, ZENG Zi-xian. Establishment of Genetic Transformation and Gene Editing System for a Potato Cultivar Chuanyu 50 [J]. Biotechnology Bulletin, 2025, 41(4): 88-97. |
| [6] | ZHANG Huan-huan, MU Xiao-ya, ZHOU Jing-yi, LYU Gao-pei, XIAO Nan, LI Min, HAO Yao-shan, WU Shen-jie. An Efficient Genetic Transformation System for High-frequency Embryogenic Broomcorn Millet Line [J]. Biotechnology Bulletin, 2025, 41(10): 164-174. |
| [7] | WANG Bi-cheng, JING Hai-qing, WAN Kun, ZHANG Ying-ying, DING Jia-hao, LI Run-zhi, XUE Jin-ai, ZHANG Hai-ping. Identification of Soybean BCAT Gene Family and Functional Analysis of GmBCAT3 in Soybean Responses to Drought Stress [J]. Biotechnology Bulletin, 2025, 41(10): 196-209. |
| [8] | LI Zhan-qian, LI Chen, LI Shu-ting, MA Ju-hua, JING Hai-qing, SUN Yan, ZHOU Ya-li, XUE Jin-ai, LI Run-zhi. Genome-wide Identification of the ME Family in Cyperus esculentusis and Functional Analysis of CeNAD-ME2 [J]. Biotechnology Bulletin, 2025, 41(10): 210-221. |
| [9] | SONG Qian-na, DUAN Yong-hong, FENG Rui-yun. Establishment of CRISPR/Cas9-mediated Highly Efficient Gene Editing System in Microtubers of Potatoes [J]. Biotechnology Bulletin, 2024, 40(9): 33-41. |
| [10] | ZHANG Mei-yu, ZHAO Yu-bin, WANG Ling-yun, SONG Yuan-da, ZHAO Xin-he, REN Xiao-jie. Research Progress in the Production of Functional Fatty Acid DHA by Microalga Thraustochytrids [J]. Biotechnology Bulletin, 2024, 40(6): 81-94. |
| [11] | MEI Xian-jun, SONG Hui-yang, LI Jing-hao, MEI Chao, SONG Qian-na, FENG Rui-yun, CHEN Xi-ming. Cloning and Expression Analysis of StDof5 Gene in Potato [J]. Biotechnology Bulletin, 2024, 40(3): 181-192. |
| [12] | HUA Xuan, TIAN Bo-wen, ZHOU Xin-tong, JIANG Zi-han, WANG Shi-qi, HUANG Qian-hui, ZHANG Jian, CHEN Yan-hong. Cloning SmERF B3-45 from Salix matsudana and Functional Analysis on Its Tolerance to Salt [J]. Biotechnology Bulletin, 2024, 40(12): 124-135. |
| [13] | HUANG Wen-li, LI Xiang-xiang, ZHOU Wen-ting, LUO Sha, YAO Wei-jia, MA Jie, ZHANG Fen, SHEN Yu-sen, GU Hong-hui, WANG Jian-sheng, SUN Bo. Targeted Editing of BoZDS in Broccoli by CRISPR/Cas9 Technology [J]. Biotechnology Bulletin, 2023, 39(2): 80-87. |
| [14] | ZHOU Jia-yan, ZOU Jian, CHEN Wei-ying, WU Yi-chao, CHEN Xi-tong, WANG Qian, ZENG Wen-jing, HU Nan, YANG Jun. Construction of Multi-gene Interference System for Plant and Analysis of Its Application Efficiency [J]. Biotechnology Bulletin, 2023, 39(1): 115-126. |
| [15] | SUN Wei, ZHANG Yan, WANG Yu-han, XU Hui, XU Xiao-rong, JU Zhi-gang. Cloning of Rd3GT1 in Rhododendron delavayi and Its Effect on Flower Color Formation of Petunia hybrida [J]. Biotechnology Bulletin, 2022, 38(9): 198-206. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||