








Biotechnology Bulletin ›› 2026, Vol. 42 ›› Issue (1): 184-197.doi: 10.13560/j.cnki.biotech.bull.1985.2025-0504
Previous Articles Next Articles
REN Yun-er( ), WU Guo-qiang(
), WU Guo-qiang( ), CHENG Bin, WEI Ming
), CHENG Bin, WEI Ming
Received:2025-05-16
Online:2026-01-26
Published:2026-02-04
Contact:
WU Guo-qiang
E-mail:954466727@qq.com;gqwu@lut.edu.cn
REN Yun-er, WU Guo-qiang, CHENG Bin, WEI Ming. Genome-wide Identification of the BvATGs Genes Family in Sugar Beet (Beta vulgaris L.) and Analysis of Their Expression Pattern under Salt Stress[J]. Biotechnology Bulletin, 2026, 42(1): 184-197.
| 基因名称 Gene name | 上游引物 Forward primer sequence (5′‒3′) | 下游引物 Reverse primer sequence (5′‒3′) |
|---|---|---|
| BvACTIN | ACTGGTATTGTGCTTGACTC | ATGAGATAATCAGTGAGATC |
| BvATG1e | CAATCTGTCTGGCTTATTCGATG | CGCTCGAAGTTTAGCAACCC |
| BvATG1h | TTTGCCGGAACACCTTAGACA | CCTTAGCACCATCATATCCCT |
| BvATG3 | CCATGCTCGCAAAACGGTA | GTCAACTTCTGGCTCAACCC |
| BvATG4 | TCGTCGTCAACTAGGGCTTC | CTCCTCCGAACGATGCTCA |
| BvATG6a-1 | GTCTCCCTAAAGTTCCGGTTG | TTTACAGGCCCAAACAGGT |
| BvATG8b | CCTACTGGAGCAATCATGTCT | CTCCACAGCAAGATACCCGAA |
| BvATG8c | GCTCCAACATCCTCTCGAAC | TGCCCCACAGTTAAATCAGC |
| BvATG8d | TCCTGAGAAAGCCATATTCGT | TAGCAATGCACCTCAATCCAA |
| BvATG8e | TGCCAAGTATCCTGATCGAGT | ATATAAATAGAGCTTTGCCCGGAG |
| BvATG10 | CCTCTCTTCACTTCCCACGA | TTGCAGGATCAATACGCTCT |
| BvATG16-2 | CAGTGCAGCTTATGATCGAAC | CAAAGCCGGAGATTCCCAT |
| BvATG20c | TCCCGAGATTTCCACTACTGC | AAGGATCGAAACATAGCGTCA |
Table 1 Primer sequences for RT-qPCR in this study
| 基因名称 Gene name | 上游引物 Forward primer sequence (5′‒3′) | 下游引物 Reverse primer sequence (5′‒3′) |
|---|---|---|
| BvACTIN | ACTGGTATTGTGCTTGACTC | ATGAGATAATCAGTGAGATC |
| BvATG1e | CAATCTGTCTGGCTTATTCGATG | CGCTCGAAGTTTAGCAACCC |
| BvATG1h | TTTGCCGGAACACCTTAGACA | CCTTAGCACCATCATATCCCT |
| BvATG3 | CCATGCTCGCAAAACGGTA | GTCAACTTCTGGCTCAACCC |
| BvATG4 | TCGTCGTCAACTAGGGCTTC | CTCCTCCGAACGATGCTCA |
| BvATG6a-1 | GTCTCCCTAAAGTTCCGGTTG | TTTACAGGCCCAAACAGGT |
| BvATG8b | CCTACTGGAGCAATCATGTCT | CTCCACAGCAAGATACCCGAA |
| BvATG8c | GCTCCAACATCCTCTCGAAC | TGCCCCACAGTTAAATCAGC |
| BvATG8d | TCCTGAGAAAGCCATATTCGT | TAGCAATGCACCTCAATCCAA |
| BvATG8e | TGCCAAGTATCCTGATCGAGT | ATATAAATAGAGCTTTGCCCGGAG |
| BvATG10 | CCTCTCTTCACTTCCCACGA | TTGCAGGATCAATACGCTCT |
| BvATG16-2 | CAGTGCAGCTTATGATCGAAC | CAAAGCCGGAGATTCCCAT |
| BvATG20c | TCCCGAGATTTCCACTACTGC | AAGGATCGAAACATAGCGTCA |
| 引物用途 Primer purpose | 引物名称 Primer name | 引物序列 Primer sequence (5′‒3′) |
|---|---|---|
基因克隆 Gene clone | BvATG4-F-Xho I | GAGGACACGCTCGAGATGAAGAACTTGTTTGAGAGTGCTGG |
| BvATG4-R-Sma I | CTTTGTAATCCCCGGGGAGGAGTTGCCATTCGTCCTCTTG | |
| BvATG6a-1-F-Xho I | TTTGGAGAGGACACGCTCGAGATGAAGGAGAAGCAGCTTG | |
| BvATG6a-1-R-Sma I | TCATCTTTGTAATCCCCGGGTTGACCAGATGGAAATCGAG | |
转基因拟南芥PCR检测 Transgenic A. thaliana PCR detection | 35S-F | GACGCACAATCCCACTATCC |
| BvATG4-R | GAGGAGTTGCCATTCGTCCTCTTG | |
| BvATG6a-1-R | TTGACCAGATGGAAATCGAG | |
内参基因 Refenrence gene | AtUBQ10-F | AGATCCAGGACAAGGAAGGTATTC |
| AtUBQ10-R | CGCAGGACCAAGTGAAGAGTAG | |
亚细胞定位 Subcellular localization | BvATG4-F-Hind III | GGACAGCCCAAGCTTATGAAGAACTTGTTTGAGAGTGCTGG |
| BvATG4-R-Nco I | TTGCTCACCATGGATCCGAGGAGTTGCCATTCGTCCTCTTG | |
| BvATG6a-1-F-Hind III | CATGGAGGCCAGTGAATTCATGAAGGAGAAGCAGCTTGATG | |
| BvATG6a-1-R-Nco I | TCGAGCTCGATGGATCCTTGACCAGATGGAAATCGAGAATC |
Table 2 Primers used in the study
| 引物用途 Primer purpose | 引物名称 Primer name | 引物序列 Primer sequence (5′‒3′) |
|---|---|---|
基因克隆 Gene clone | BvATG4-F-Xho I | GAGGACACGCTCGAGATGAAGAACTTGTTTGAGAGTGCTGG |
| BvATG4-R-Sma I | CTTTGTAATCCCCGGGGAGGAGTTGCCATTCGTCCTCTTG | |
| BvATG6a-1-F-Xho I | TTTGGAGAGGACACGCTCGAGATGAAGGAGAAGCAGCTTG | |
| BvATG6a-1-R-Sma I | TCATCTTTGTAATCCCCGGGTTGACCAGATGGAAATCGAG | |
转基因拟南芥PCR检测 Transgenic A. thaliana PCR detection | 35S-F | GACGCACAATCCCACTATCC |
| BvATG4-R | GAGGAGTTGCCATTCGTCCTCTTG | |
| BvATG6a-1-R | TTGACCAGATGGAAATCGAG | |
内参基因 Refenrence gene | AtUBQ10-F | AGATCCAGGACAAGGAAGGTATTC |
| AtUBQ10-R | CGCAGGACCAAGTGAAGAGTAG | |
亚细胞定位 Subcellular localization | BvATG4-F-Hind III | GGACAGCCCAAGCTTATGAAGAACTTGTTTGAGAGTGCTGG |
| BvATG4-R-Nco I | TTGCTCACCATGGATCCGAGGAGTTGCCATTCGTCCTCTTG | |
| BvATG6a-1-F-Hind III | CATGGAGGCCAGTGAATTCATGAAGGAGAAGCAGCTTGATG | |
| BvATG6a-1-R-Nco I | TCGAGCTCGATGGATCCTTGACCAGATGGAAATCGAGAATC |
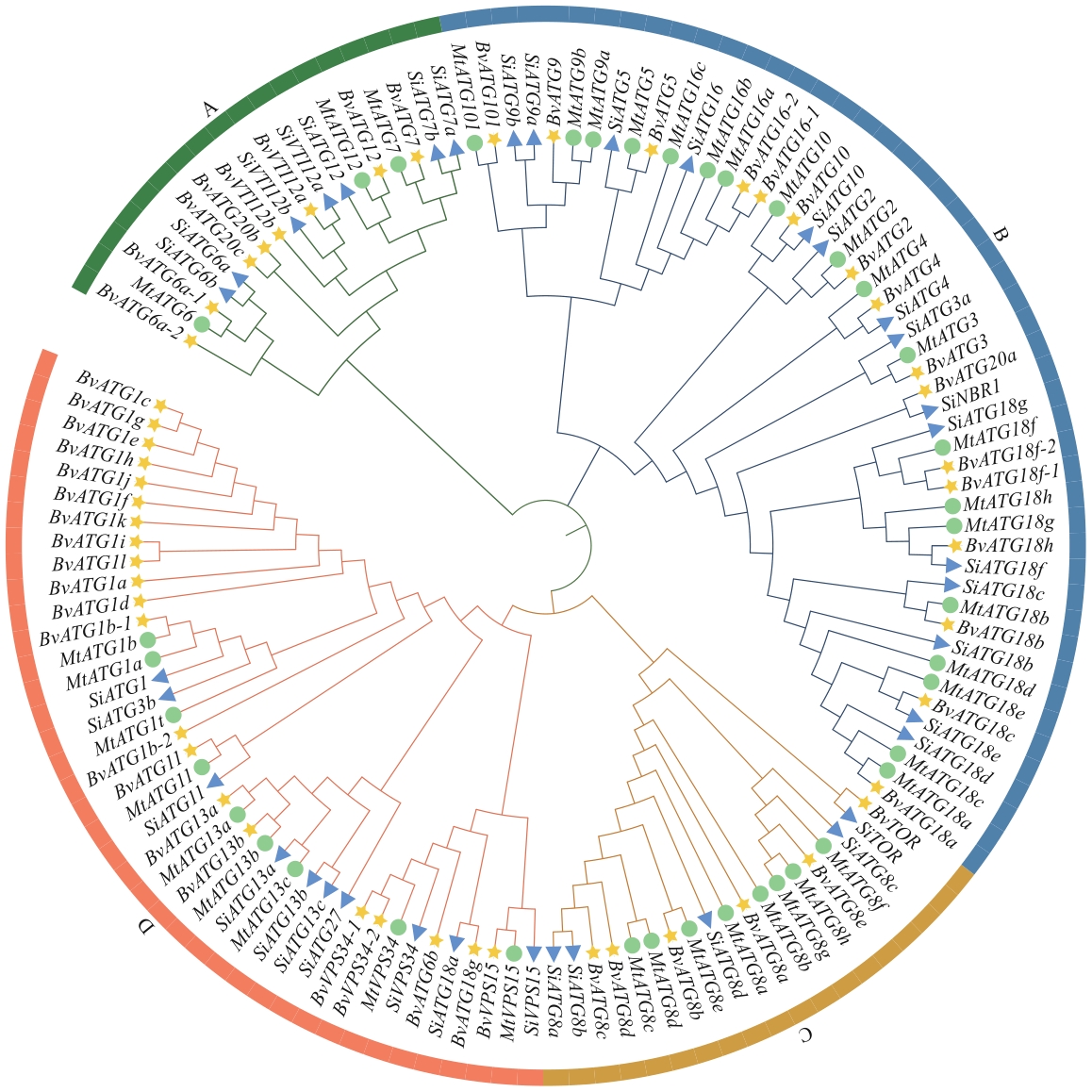
Fig. 2 Phylogenetic analysis of ATGs in higher plantsThe yellow pentagons indicate sugar beet (B. vulgaris), the green circles indicate M. truncatula and the blue triangles indicate S. italica. The name and accession number of ATGs are shown in the Supplementary Table S2
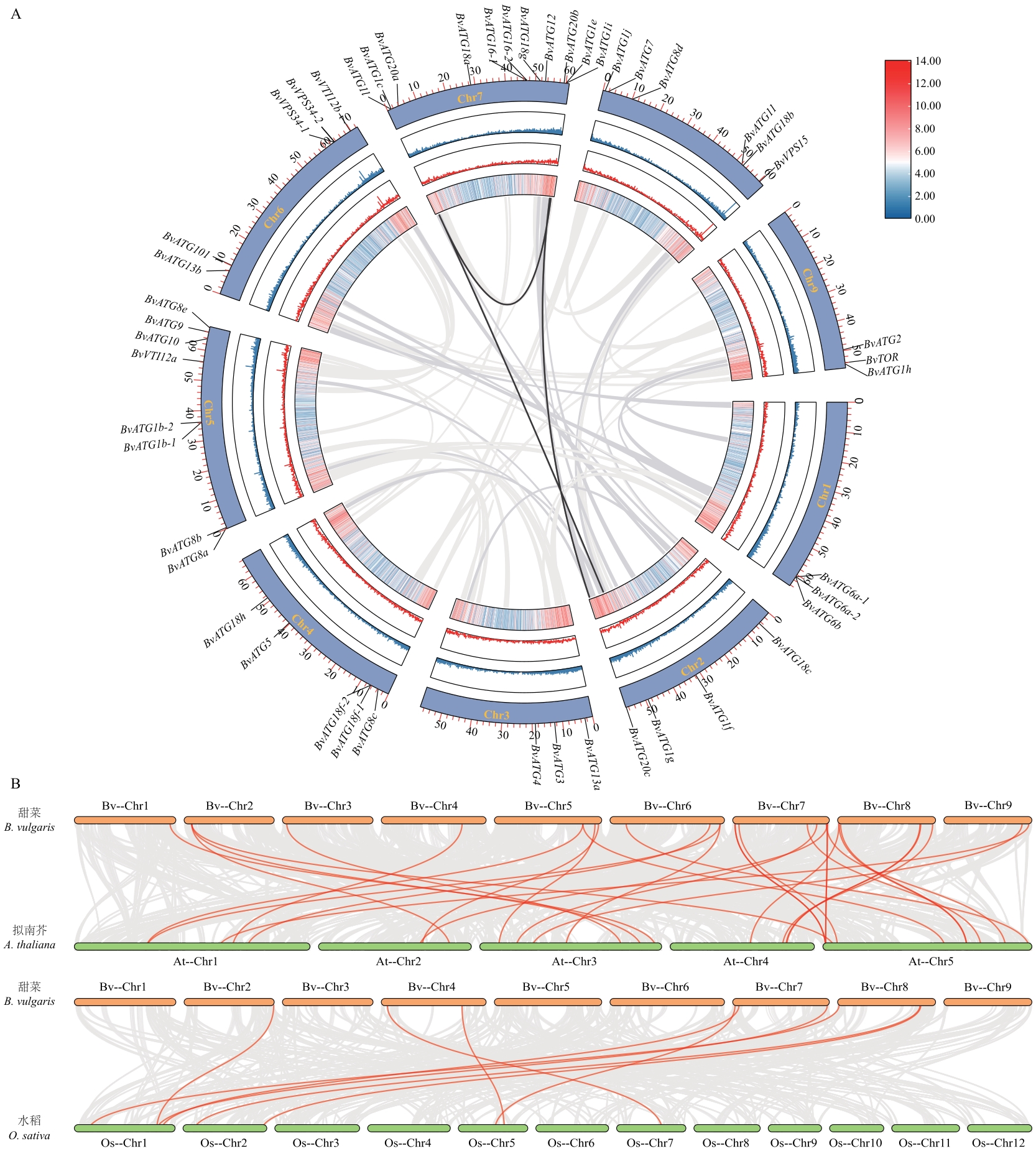
Fig. 5 Collinearity analysis of BvATGs gene family in sugar beetA: ATGs gene family intraspecies collinearity analysis. B: Synteny analysis of ATG genes between sugar beet (B. vulgaris), rice (O. sativa) and A. thaliana

Fig. 6 Relative expressions of BvATGs in the shoots and roots of sugar beet plants treated with different concentrations of NaCl for 12 h (A) and 24 h (B)Data are normalized to BvACTIN expression. Vertical bars indicate standard error (SE) (n = 3). Different lowercase letters on the bars indicate significant difference at P < 0.05 level. The same below
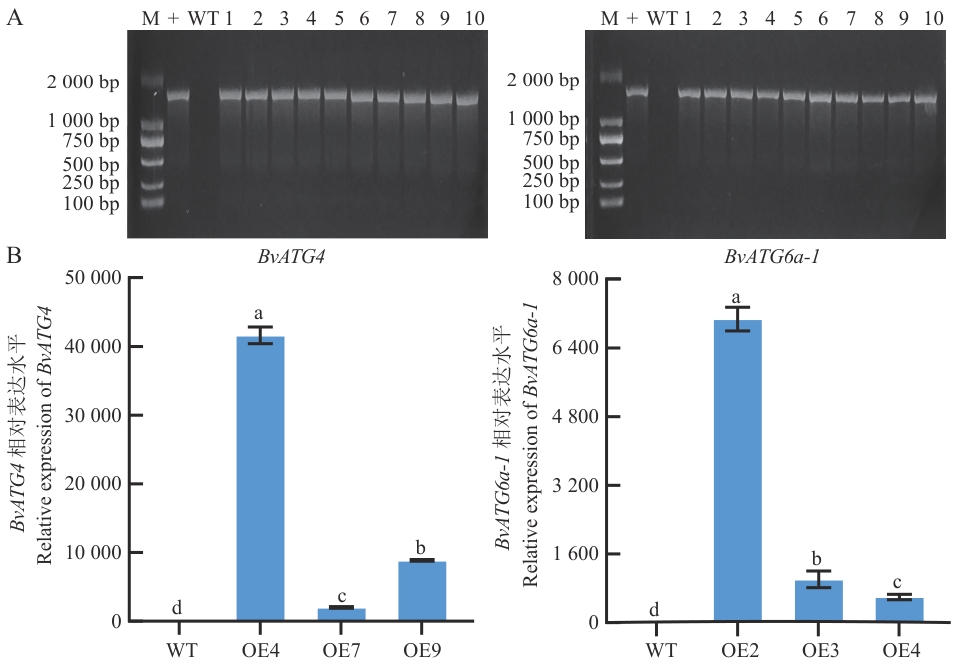
Fig. 8 Identification of BvATG4 and BvATG6a-1 transgenic A. thalianaA: Genomic DNA PCR detection of BvATG4 and BvATG6a-1 transgenic A. thaliana. B: RT-qPCR detection of BvATG4 and BvATG6a-1 transgenic A. thaliana. M: DL2000 marker for molecular weight; + : pEG-BvATG4 and pEG-BvATG6a-1 plasmid. WT: Wild-type A. thaliana; 1-10: BvATG4 and BvATG6a-1 transgenic A. thaliana; OE4, OE7, OE9: BvATG4 transgenic A. thaliana; OE2, OE3, OE4: BvATG6a-1 transgenic A. thaliana
| [1] | Liang XY, Li JF, Yang YQ, et al. Designing salt stress-resilient crops: Current progress and future challenges [J]. J Integr Plant Biol, 2024, 66(3): 303-329. |
| [2] | Zhou HP, Shi HF, Yang YQ, et al. Insights into plant salt stress signaling and tolerance [J]. J Genet Genom, 2024, 51(1): 16-34. |
| [3] | Ashraf M, Munns R. Evolution of approaches to increase the salt tolerance of crops [J]. Crit Rev Plant Sci, 2022, 41(2): 128-160. |
| [4] | Azmat MA, Zaheer M, Shaban M, et al. Autophagy: a new avenue and biochemical mechanisms to mitigate the climate change [J]. Scientifica, 2024, 2024(1): 9908323. |
| [5] | Yagyu M, Yoshimoto K. New insights into plant autophagy: molecular mechanisms and roles in development and stress responses [J]. J Exp Bot, 2024, 75(5): 1234-1251. |
| [6] | Sedaghatmehr M, Balazadeh S. Autophagy: a key player in the recovery of plants from heat stress [J]. J Exp Bot, 2024, 75(8): 2246-2255. |
| [7] | Suttangkakul A, Li FQ, Chung T, et al. The ATG1/ATG13 protein kinase complex is both a regulator and a target of autophagic recycling in Arabidopsis [J]. Plant Cell, 2011, 23(10): 3761-3779. |
| [8] | Tsukada M, Ohsumi Y. Isolation and characterization of autophagy-defective mutants of Saccharomyces cerevisiae [J]. FEBS Lett, 1993, 333(1/2): 169-174. |
| [9] | Huang L, Wen X, Jin L, et al. HOOKLESS1 acetylates AUTOPHAGY-RELATED PROTEIN18a to promote autophagy during nutrient starvation in Arabidopsis [J]. Plant Cell, 2023, 36(1): 136-157. |
| [10] | Kwon SI, Park OK. Autophagy in plants [J]. J Plant Biol, 2008, 51(5): 313-320. |
| [11] | Han SJ, Yu BJ, Wang Y, et al. Role of plant autophagy in stress response [J]. Protein Cell, 2011, 2(10): 784-791. |
| [12] | Yadav M, Saxena G, Verma RK, et al. Genome-wide identification and expression analysis of autophagy-related genes (ATG) in Gossypium spp. reveals their crucial role in stress tolerance [J]. S Afr N J Bot, 2024, 167: 82-93. |
| [13] | Liu H, Liu Y, Zhang YT, et al. Genome-wide identification and expression analysis of autophagy-related genes (ATGs), revealing ATG8a and ATG18b participating in drought stress in Phoebe bournei [J]. Environ Exp Bot, 2024, 228: 106012. |
| [14] | Wang Y, Ban QY, Liu TJ, et al. Genome-wide identification and expression analysis of autophagy-related genes in eggplant (Solanum melongena L.) [J]. Sci Hortic, 2024, 330: 113085. |
| [15] | Wang Y, Sun X, Zhang ZW, et al. Genome-wide identification and characterization of the PbrATG family in Pyrus bretschneideri and functional analysis of PbrATG1a in response to Botryosphaeria dothidea [J]. Hortic Plant J, 2024, 10(2): 327-340. |
| [16] | Shi Y, Wu Y, Li ML, et al. Genome-wide identification and analysis of autophagy-related (ATG) genes in Lycium ruthenicum Murray reveals their crucial roles in salt stress tolerance [J]. Plant Sci, 2025, 352: 112371. |
| [17] | Luo KS, Li JH, Lu M, et al. Genome-wide identification and expression analysis of Rosa roxburghii autophagy-related genes in response to top-rot disease [J]. Biomolecules, 2023, 13(3): 556. |
| [18] | Petersen M, Avin-Wittenberg T, Bassham DC, et al. Autophagy in plants [J]. Autophagy Rep, 2024, 3(1): 2395731. |
| [19] | Luo LM, Zhang PP, Zhu RH, et al. Autophagy is rapidly induced by salt stress and is required for salt tolerance in Arabidopsis [J]. Front Plant Sci, 2017, 8: 1459. |
| [20] | Liu ML, Ma L, Tang Y, et al. Maize autophagy-related protein ZmATG3 confers tolerance to multiple abiotic stresses [J]. Plants, 2024, 13(12): 1637. |
| [21] | Su WL, Bao Y, Lu YY, et al. Poplar autophagy receptor NBR1 enhances salt stress tolerance by regulating selective autophagy and antioxidant system [J]. Front Plant Sci, 2020, 11: 568411. |
| [22] | Yu XQ, Su WL, Zhang H, et al. Genome-wide analysis of autophagy-related gene family and PagATG18a enhances salt tolerance by regulating ROS homeostasis in poplar [J]. Int J Biol Macromol, 2023, 224: 1524-1540. |
| [23] | Yang J, Qiu LN, Mei QL, et al. MdHB7-like positively modulates apple salt tolerance by promoting autophagic activity and Na+ efflux [J]. Plant J, 2023, 116(3): 669-689. |
| [24] | Li NN, Wang W, Guo XT, et al. BvBZR1 improves parenchyma cell development and sucrose accumulation in sugar beet (Beta vulgaris L.) taproot [J]. Front Plant Sci, 2025, 16: 1495161. |
| [25] | Obama R, Kumagai K, Maruyama H, et al. Effect of different light conditions on the growth response of sugar beet to NaCl [J]. Soil Sci Plant Nutr, 2025, 71(2): 113-121. |
| [26] | Yang L, Ma CQ, Wang LL, et al. Salt stress induced proteome and transcriptome changes in sugar beet monosomic addition line M14 [J]. J Plant Physiol, 2012, 169(9): 839-850. |
| [27] | Sánchez-Sastre LF, Alte da Veiga NMS, Ruiz-Potosme NM, et al. Sugar beet agronomic performance evolution in NW Spain in future scenarios of climate change [J]. Agronomy, 2020, 10(1): 91. |
| [28] | Lin XC, Song BQ, Adil MF, et al. Response of the rhizospheric soil microbial community of sugar beet to nitrogen application: a case of black soil in Northeast China [J]. Appl Soil Ecol, 2023, 191: 105050. |
| [29] | Wu GQ, Feng RJ, Liang N, et al. Sodium chloride stimulates growth and alleviates sorbitol-induced osmotic stress in sugar beet seedlings [J]. Plant Growth Regul, 2015, 75(1): 307-316. |
| [30] | Dohm JC, Minoche AE, Holtgräwe D, et al. The genome of the recently domesticated crop plant sugar beet (Beta vulgaris) [J]. Nature, 2014, 505(7484): 546-549. |
| [31] | Hu YD, Ren PP, Wei M, et al. Genome-wide identification of shaker K+ channel gene family in sugar beet (Beta vulgaris L.) and function of BvSKOR in response to salt and drought stresses [J]. Environ Exp Bot, 2024, 228: 106034. |
| [32] | Zhang JL, Wu GQ, Ma BT, et al. Genome-wide identification of the BvHSFs gene family and their expression in sugar beet (Beta vulgaris L.) under salt stress [J]. Sugar Tech, 2024, 26(5): 1463-1476. |
| [33] | Zhang XM, Wu GQ, Wei M. Genome-wide identification of sugar beet (Beta vulgaris L.) MAPKKKs gene family and their expression in response to salt stress [J]. Sugar Tech, 2024, 26(5): 1337-1349. |
| [34] | Ionascu A, Al Ecovoiu A, Chifiriuc MC, et al. qDATA - an R application implementing a practical framework for analyzing quantitative real-time PCR data [J]. Biotechniques, 2024, 76(12): 559-573. |
| [35] | Yang MK, Wang LP, Chen CM, et al. Genome-wide analysis of autophagy-related genes in Medicago truncatula highlights their roles in seed development and response to drought stress [J]. Sci Rep, 2021, 11(1): 22933. |
| [36] | Taneja M, Tyagi S, Sharma S, et al. Ca2+ cation antiporters (CaCA): identification, characterization and expression profiling in bread wheat (Triticum aestivum L.) [J]. Front Plant Sci, 2016, 7: 1775. |
| [37] | Hu YH, Lehrach H, Janitz M. Comparative analysis of an experimental subcellular protein localization assay and in silico prediction methods [J]. J Mol Histol, 2009, 40(5/6): 343-352. |
| [38] | Qiao X, Li QH, Yin H, et al. Gene duplication and evolution in recurring polyploidization-diploidization cycles in plants [J]. Genome Biol, 2019, 20(1): 38. |
| [39] | Qiao X, Yin H, Li LT, et al. Different modes of gene duplication show divergent evolutionary patterns and contribute differently to the expansion of gene families involved in important fruit traits in pear (Pyrus bretschneideri) [J]. Front Plant Sci, 2018, 9: 161. |
| [40] | Hu YF, Zhang M, Yin FR, et al. Genome-wide identification and expression analysis of BrATGs and their different roles in response to abiotic stresses in Chinese cabbage [J]. Agronomy, 2022, 12(12): 2976. |
| [41] | Sheshadri SA, Nishanth MJ, Simon B. Stress-mediated cis-element transcription factor interactions interconnecting primary and specialized metabolism in planta [J]. Front Plant Sci, 2016, 7: 1725. |
| [42] | Wu XY, Xia M, Su P, et al. MYB transcription factors in plants: a comprehensive review of their discovery, structure, classification, functional diversity and regulatory mechanism [J]. Int J Biol Macromol, 2024, 282(Pt 2): 136652. |
| [43] | Dong P, Xiong FJ, Que YM, et al. Expression profiling and functional analysis reveals that TOR is a key player in regulating photosynthesis and phytohormone signaling pathways in Arabidopsis [J]. Front Plant Sci, 2015, 6: 677. |
| [44] | Fang Y, Wang S, Wu HL, et al. Genome-wide identification of ATG gene family members in Fagopyrum tataricum and their expression during stress responses [J]. Int J Mol Sci, 2022, 23(23): 14845. |
| [45] | Zhai YF, Guo M, Wang H, et al. Autophagy, a conserved mechanism for protein degradation, responds to heat, and other abiotic stresses in Capsicum annuum L [J]. Front Plant Sci, 2016, 7: 131. |
| [46] | Fu XZ, Zhou X, Xu YY, et al. Comprehensive analysis of autophagy-related genes in sweet orange (Citrus sinensis) highlights their roles in response to abiotic stresses [J]. Int J Mol Sci, 2020, 21(8): 2699. |
| [47] | Wang JR, Zhou GQ, Huang WX, et al. Autophagy-related gene PlATG6a is involved in mycelial growth, asexual reproduction and tolerance to salt and oxidative stresses in Peronophythora litchii [J]. Int J Mol Sci, 2022, 23(3): 1839. |
| [1] | ZENG Ting, ZHANG Lan, LUO Rui. Functional Analysis of the Transcription Factor MpR2R3-MYB17 in Regulating Gemma Development in Marchantia polymorpha L. [J]. Biotechnology Bulletin, 2026, 42(1): 208-217. |
| [2] | LYU Cheng-cong, HENG Meng, CHEN Si-qi, JIN Xue-hua. Cloning and Functional Analysis of ZhGSTF Related to Anthocyanin Transport Zantedeschia hybrida [J]. Biotechnology Bulletin, 2026, 42(1): 161-169. |
| [3] | YANG Dan, JIN Ya-rong, MAO Chun-li, WANG Bi-xian, ZHANG Ya-ning, YANG Zhi-yi, ZHOU Zhi-yao, YANG Rui-ming, FAN Heng-rui, HUANG Lin-kai, YAN Hai-dong. Identification and Expression Analysis of C2H2 Gene Family in Cenchrus purpureus [J]. Biotechnology Bulletin, 2026, 42(1): 251-261. |
| [4] | YANG Yue-qin, XING Ying, ZHONG Zi-he, TIAN Wei-jun, YANG Xue-qing, WANG Jian-xu. Expression and Functional Analysis of OsMATE34 in Rice under Mercury Stress [J]. Biotechnology Bulletin, 2026, 42(1): 86-94. |
| [5] | YANG Juan, FENG Hui, JI Nai-zhe, SUN Li-ping, WANG Yun, ZHANG Jia-nan, ZHAO Shi-wei. Cloning and Functional Analysis of AP2/ERF Transcription Factors RcERF4 and RcRAP2-12 in Rose [J]. Biotechnology Bulletin, 2026, 42(1): 150-160. |
| [6] | ZHANG Yue, DAI Yue-hua, ZHANG Ying-ying, LI Ao-hui, LI Chu-hui, XUE Jin-ai, QIN Hui-bin, CHEN Yan, NIE Meng-en, ZHANG Hai-ping. Cloning and Functional Analysis of the Soybean Enoyl-CoA Reductase ECR14 Gene [J]. Biotechnology Bulletin, 2026, 42(1): 95-104. |
| [7] | WU Cui-cui, CHEN Deng-ke, LAN Gang, XIA Zhi, LI Peng-bo. Bioinformatics Analysis of Peanut Transcription Factor AhHDZ70 and Its Tolerances to Salt and Drought [J]. Biotechnology Bulletin, 2026, 42(1): 198-207. |
| [8] | CHEN Jing-huan, FANG Guo-nan, ZHU Wen-hao, YE Guang-ji, SU Wang, HE Miao-miao, YANG Sheng-long, ZHOU Yun. Starch Characterization and Related Gene Expression Analysis of Potato Germplasm Resources [J]. Biotechnology Bulletin, 2026, 42(1): 170-183. |
| [9] | DONG Xiang-xiang, MIAO Bai-ling, XU He-juan, CHEN Juan-juan, LI Liang-jie, GONG Shou-fu, ZHU Qing-song. Bioinformatics Analysis and Flowering Regulation Function of FveBBX20 Gene in Woodland Strawberry [J]. Biotechnology Bulletin, 2025, 41(9): 115-123. |
| [10] | LI Shan, MA Deng-hui, MA Hong-yi, YAO Wen-kong, YIN Xiao. Identification and Expression Analysis of SKP1 Gene Family in Grapevine (Vitis vinifera L.) [J]. Biotechnology Bulletin, 2025, 41(9): 147-158. |
| [11] | HUANG Guo-dong, DENG Yu-xing, CHENG Hong-wei, DAN Yan-nan, ZHOU Hui-wen, WU Lan-hua. Genome-wide Identification and Expression Analysis of the ZIP Gene Family in Soybean [J]. Biotechnology Bulletin, 2025, 41(9): 71-81. |
| [12] | GONG Hui-ling, XING Yu-jie, MA Jun-xian, CAI Xia, FENG Zai-ping. Identification of Laccase (LAC) Gene Family in Potato (Solanum tuberosum L.) and Its Expression Analysis under Salt Stresses [J]. Biotechnology Bulletin, 2025, 41(9): 82-93. |
| [13] | LIU Jia-li, SONG Jing-rong, ZHAO Wen-yu, ZHANG Xin-yuan, ZHAO Zi-yang, CAO Yi-bo, ZHANG Ling-yun. Identification of the R2R3-MYB Gene and Expression Analysis of Flavonoid Regulatory Genes in Blueberry [J]. Biotechnology Bulletin, 2025, 41(9): 124-138. |
| [14] | CHENG Ting-ting, LIU Jun, WANG Li-li, LIAN Cong-long, WEI Wen-jun, GUO Hui, WU Yao-lin, YANG Jing-fan, LAN Jin-xu, CHEN Sui-qing. Genome-wide Identification of the Chalcone Isomerase Gene Family in Eucommia ulmoides and Analysis of Their Expression Patterns [J]. Biotechnology Bulletin, 2025, 41(9): 242-255. |
| [15] | XU Xiao-ping, YANG Cheng-long, HE Xing, GUO Wen-jie, WU Jian, FANG Shao-zhong. Cloning of the LoAPS1 and Its Function Analysis during the Process of Dormancy Release in Lilium [J]. Biotechnology Bulletin, 2025, 41(9): 195-206. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||