








Biotechnology Bulletin ›› 2026, Vol. 42 ›› Issue (1): 198-207.doi: 10.13560/j.cnki.biotech.bull.1985.2025-0901
Previous Articles Next Articles
WU Cui-cui1( ), CHEN Deng-ke1,2(
), CHEN Deng-ke1,2( ), LAN Gang1, XIA Zhi1, LI Peng-bo1(
), LAN Gang1, XIA Zhi1, LI Peng-bo1( )
)
Received:2025-08-20
Online:2026-01-26
Published:2026-02-04
Contact:
LI Peng-bo
E-mail:wucuicui19821021@126.com;lipengbo@sxau.edu.cn
WU Cui-cui, CHEN Deng-ke, LAN Gang, XIA Zhi, LI Peng-bo. Bioinformatics Analysis of Peanut Transcription Factor AhHDZ70 and Its Tolerances to Salt and Drought[J]. Biotechnology Bulletin, 2026, 42(1): 198-207.
| 基因名称 Gene name | 正向引物 Forward primer (5′‒3′) | 反向引物 Reverse primer (5′‒3′) | 实验类型 Type of experiment |
|---|---|---|---|
| AhHDZ70 | TTGGAGAGAACACGGGGGACGAATTCATGGAATATGCAACATATTCATCAGC | GCCTGCAGGTCGACTCTAGAGGATCCGGACCAAAAGTCCCACCATTG | 过表达实验、亚定位实验、过表达PCR鉴定 |
| β-Actin | CAGGATTTGCCGGTGATGATG | TCTGTTGGCCTTCGGGTTGAG | 内参 |
| AhHDZ70 | AGATGAGTTTGGGTTTGAGT | GAGGTAGAGGCTTCTGGAT | RT-qPCR |
Table 1 Primers used in the study
| 基因名称 Gene name | 正向引物 Forward primer (5′‒3′) | 反向引物 Reverse primer (5′‒3′) | 实验类型 Type of experiment |
|---|---|---|---|
| AhHDZ70 | TTGGAGAGAACACGGGGGACGAATTCATGGAATATGCAACATATTCATCAGC | GCCTGCAGGTCGACTCTAGAGGATCCGGACCAAAAGTCCCACCATTG | 过表达实验、亚定位实验、过表达PCR鉴定 |
| β-Actin | CAGGATTTGCCGGTGATGATG | TCTGTTGGCCTTCGGGTTGAG | 内参 |
| AhHDZ70 | AGATGAGTTTGGGTTTGAGT | GAGGTAGAGGCTTCTGGAT | RT-qPCR |
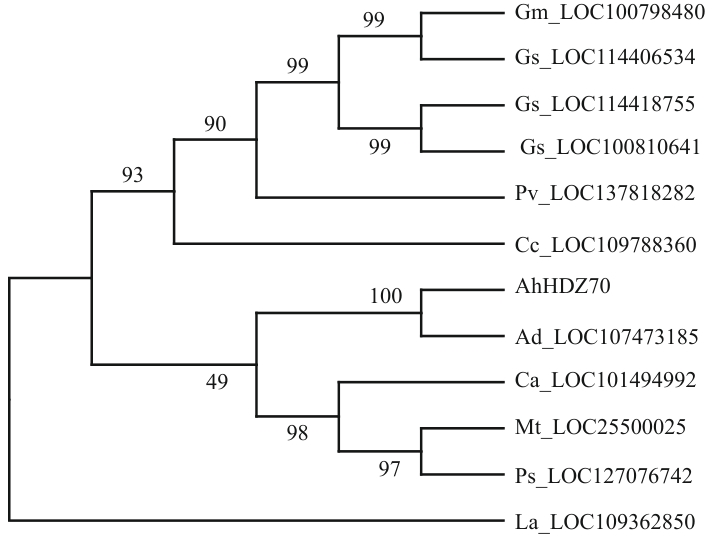
Fig. 1 Phylogenetic tree analysis of homologous proteins between AhHDZ70 and other speciesAh: Arachis hypogaea; Ad: Arachis duranensis; Ca: Cicer arietinum; Cc: Cajanus cajan; Gm: Glycine max; Gs: Glycine soja; La: Lupinus angustifolius; Mt: Medicago truncatula; Ps: Pisum sativum; Pv: Phaseolus vulgaris
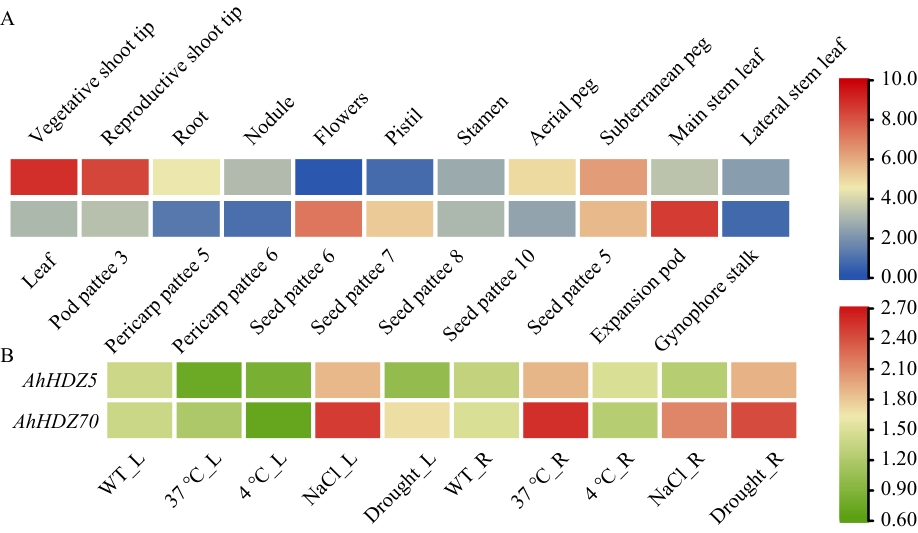
Fig. 3 Analysis of AhHDZ70 expression patterns in tissues, organs, and abiotic stressA: Expression patterns of AhHDZ70 in 22 tissues and organs. B: Expression patterns of AhHDZ70 under abiotic stress. L: Leaf. R: Root
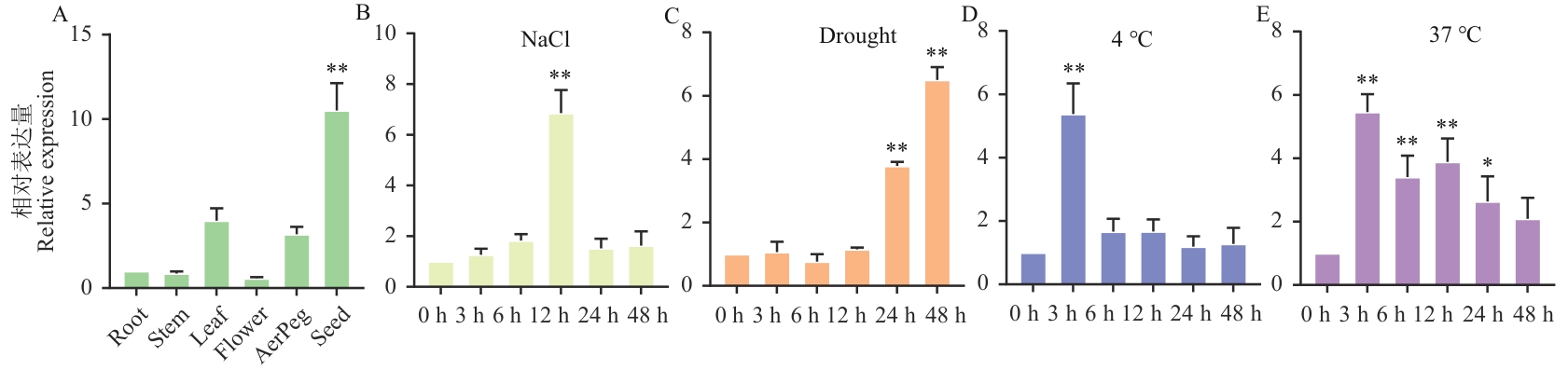
Fig. 4 RT-qPCR identification of AhHDZ70 in different tissues and organs of peanut under abiotic stressA: RT-qPCR analysis of AhHDZ70 in different tissues and organs of peanuts. B-E: RT-qPCR analysis of AhHDZ70 under salt, drought, low temperature and high temperature stress. The difference significance was analyzed by two-tailed t-test. Error bars indicate mean ±SD, *P<0.05, **P<0.01,the same below
| [1] | de Carvalho Moretzsohn M, Hopkins MS, Mitchell SE, et al. Genetic diversity of peanut (Arachis hypogaea L.) and its wild relatives based on the analysis of hypervariable regions of the genome [J]. BMC Plant Biol, 2004, 4(1): 11. |
| [2] | Desmae H, Janila P, Okori P, et al. Genetics, genomics and breeding of groundnut (Arachis hypogaea L.) [J]. Plant Breed, 2019, 138(4): 425-444. |
| [3] | Zhuang WJ, Chen H, Yang M, et al. The genome of cultivated peanut provides insight into legume karyotypes, polyploid evolution and crop domestication [J]. Nat Genet, 2019, 51(5): 865-876. |
| [4] | 郑青焕, 李拴柱, 王建玉, 等. 河南省鲜食花生研究现状与发展前景 [J]. 中国种业, 2025(8): 19-23. |
| Zheng QH, Li SZ, Wang JY, et al. Research status and development prospects of fresh peanuts in Henan province [J]. China Seed Ind, 2025(8): 19-23. | |
| [5] | 李博文,赵建军. 挖掘盐碱地改造利用潜力[J].经济日报, 2024, 11, 24. |
| [6] | Wu CC, Hou BG, Wu RL, et al. Genome-wide analysis elucidates the roles of AhLBD genes in different abiotic stresses and growth and development stages in the peanut (Arachis hypogea L.) [J]. Int J Mol Sci, 2024, 25(19): 10561. |
| [7] | Chen XP, Lu Q, Liu H, et al. Sequencing of cultivated peanut, Arachis hypogaea, yields insights into genome evolution and oil improvement [J]. Mol Plant, 2019, 12(7): 920-934. |
| [8] | Bertioli DJ, Jenkins J, Clevenger J, et al. The genome sequence of segmental allotetraploid peanut Arachis hypogaea [J]. Nat Genet, 2019, 51(5): 877-884. |
| [9] | Zhao KK, Xue HZ, Li GW, et al. Pangenome analysis reveals structural variation associated with seed size and weight traits in peanut [J]. Nat Genet, 2025, 57(5): 1250-1261. |
| [10] | Lu Q, Huang L, Liu H, et al. A genomic variation map provides insights into peanut diversity in China and associations with 28 agronomic traits [J]. Nat Genet, 2024, 56(3): 530-540. |
| [11] | Hrmova M, Hussain SS. Plant transcription factors involved in drought and associated stresses [J]. Int J Mol Sci, 2021, 22(11): 5662. |
| [12] | Chen Y, Yin ML, Sun LY, et al. MYB gene family in Magnolia biondii: Identification and functional roles in waterlogging stress response [J]. Plant Physiol Biochem, 2025, 229: 110505. |
| [13] | Yu YA, Wu YX, He LY. A wheat WRKY transcription factor TaWRKY17 enhances tolerance to salt stress in transgenic Arabidopsis and wheat plant [J]. Plant Mol Biol, 2023, 113(4/5): 171-191. |
| [14] | Li L, Zhao ZZ, Li WQ, et al. The bHLH transcription factor PdbUNE12 functions as a positive regulator of the salt stress response in Populus davidiana × Populus bolleana [J]. Physiol Plant, 2025, 177(5): e70531. |
| [15] | Qiang ZQ, Zeng Z, Ma DF, et al. NAC transcription factor LpNAC22 positively regulates drought tolerance in perennial ryegrass [J]. Plant Cell Environ, 2025, 48(10): 7256-7270. |
| [16] | Chen N, Pan LJ, Yang Z, et al. A MYB-related transcription factor from peanut, AhMYB30, improves freezing and salt stress tolerance in transgenic Arabidopsis through both DREB/CBF and ABA-signaling pathways [J]. Front Plant Sci, 2023, 14: 1136626. |
| [17] | Yuan CL, Li CJ, Lu XD, et al. Comprehensive genomic characterization of NAC transcription factor family and their response to salt and drought stress in peanut [J]. BMC Plant Biol, 2020, 20(1): 454. |
| [18] | Zhao XB, Wang Q, Yan CX, et al. The bHLH transcription factor AhbHLH121 improves salt tolerance in peanut [J]. Int J Biol Macromol, 2024, 256: 128492. |
| [19] | Elhiti M, Stasolla C. Structure and function of homodomain-leucine zipper (HD-Zip) proteins [J]. Plant Signal Behav, 2009, 4(2): 86-88. |
| [20] | Shao JX, Haider I, Xiong LZ, et al. Functional analysis of the HD-Zip transcription factor genes Oshox12 and Oshox14 in rice [J]. PLoS One, 2018, 13(7): e0199248. |
| [21] | Lin ZF, Hong YG, Yin MG, et al. A tomato HD-Zip homeobox protein, LeHB-1, plays an important role in floral organogenesis and ripening [J]. Plant J, 2008, 55(2): 301-310. |
| [22] | Jiao P, Jiang ZZ, Wei XT, et al. Overexpression of the homeobox-leucine zipper protein ATHB-6 improves the drought tolerance of maize (Zea mays L.) [J]. Plant Sci, 2022, 316: 111159. |
| [23] | Ren AP, Wen TY, Xu X, et al. Cotton HD-Zip I transcription factor GhHB4-like regulates the plant response to salt stress [J]. Int J Biol Macromol, 2024, 278: 134857. |
| [24] | Possenti M, Sessa G, Alfè A, et al. HD-Zip II transcription factors control distal stem cell fate in Arabidopsis roots by linking auxin signaling to the FEZ/SOMBRERO pathway [J]. Development, 2024, 151(8): dev202586. |
| [25] | Zhao YQ, Han Q, Kang XK, et al. The HAT1 transcription factor regulates photomorphogenesis and skotomorphogenesis via phytohormone levels [J]. Plant Physiol, 2024, 197(1): kiae542. |
| [26] | Baek W, Bae Y, Lim CW, et al. Pepper homeobox abscisic acid signalling-related transcription factor 1, CaHAT1, plays a positive role in drought response [J]. Plant Cell Environ, 2023, 46(7): 2061-2077. |
| [27] | Ariel FD, Manavella PA, Dezar CA, et al. The true story of the HD-Zip family [J]. Trends Plant Sci, 2007, 12(9): 419-426. |
| [28] | Li SJ, Yu ML, Qanmber GL, et al. GhHB14_D10 and GhREV_D5, two HD-ZIP III transcription factors, play a regulatory role in cotton fiber secondary cell wall biosynthesis [J]. Plant Cell Rep, 2024, 43(3): 76. |
| [29] | Mabuchi A, Soga K, Wakabayashi K, et al. Phenotypic screening of Arabidopsis T-DNA insertion lines for cell wall mechanical properties revealed ANTHOCYANINLESS2, a cell wall-related gene [J]. J Plant Physiol, 2016, 191: 29-35. |
| [30] | Ren WB, Wu CC, Wang T, et al. GhHDZ50 regulates cotton fiber elongation in Gossypium hirsutum L. through control of fatty acid biosynthesis [J]. Plant Sci, 2025, 359: 112641. |
| [31] | Clevenger J, Chu Y, Scheffler B, et al. A developmental transcriptome map for allotetraploid Arachis hypogaea [J]. Front Plant Sci, 2016, 7: 1446. |
| [32] | Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative CT method [J]. Nat Protoc, 2008, 3(6): 1101-1108. |
| [33] | Chen CJ, Chen H, Zhang Y, et al. TBtools: an integrative toolkit developed for interactive analyses of big biological data [J]. Mol Plant, 2020, 13(8): 1194-1202. |
| [34] | Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana [J]. Plant J, 1998, 16(6): 735-743. |
| [35] | Wang Q, Wang YY, Zhang FH, et al. Genome-wide characterisation of HD-Zip transcription factors and functional analysis of PbHB24 during stone cell formation in Chinese white pear (Pyrus bretschneideri) [J]. BMC Plant Biol, 2024, 24(1): 444. |
| [36] | 刘育佼, 闫小玲, 畅欣, 等. 毛竹HD-Zip转录因子Pehox14基因的克隆及其在干旱和盐胁迫下的功能分析 [J]. 农业生物技术学报, 2025, 33(7): 1476-1489. |
| Liu YJ, Yan XL, Chang X, et al. Cloning of HD-zip transcription factor Pehox14 gene in moso bamboo (Phyllostachys edulis) and its functional analysis under drought and salt stress [J]. J Agric Biotechnol, 2025, 33(7): 1476-1489. | |
| [37] | Huang H, Wang H, Tong Y, et al. Identification and characterization of HD-Zip genes reveals their roles in stresses responses and facultative crassulacean acid metabolism in Dendrobium catenatum [J]. Sci Hortic, 2021, 285: 110058. |
| [38] | 黄丹, 彭兵阳, 张盼盼, 等. 油茶HD-Zip基因家族鉴定及其在非生物胁迫下的表达分析 [J]. 生物技术通报, 2025, 41(6): 191-207. |
| Huang D, Peng BY, Zhang PP, et al. Identification of HD-Zip gene family in Camellia oleifera and analysis of its expression under abiotic stress [J]. Biotechnol Bull, 2025, 41(6): 191-207. | |
| [39] | Li W, Dong JY, Cao MX, et al. Genome-wide identification and characterization of HD-ZIP genes in potato [J]. Gene, 2019, 697: 103-117. |
| [40] | 刘梦梦, 刘晓, 尤倩, 等. 半夏HD-Zip基因家族全基因组鉴定及表达分析 [J]. 农业生物技术学报, 2024, 32(11): 2540-2551. |
| Liu MM, Liu X, You Q, et al. Genome identification and expression analysis of HD-Zip gene family in Pinellia ternata [J]. J Agric Biotechnol, 2024, 32(11): 2540-2551. | |
| [41] | 吴翠翠, 肖水平. 陆地棉HD-Zip家族全基因组鉴定及响应非生物胁迫的表达分析 [J]. 生物技术通报, 2024, 40(2): 130-145. |
| Wu CC, Xiao SP. Genome-wide identification of HD-Zip gene family in Gossypium hirsutum L. and expression analysis in response to abiotic stress [J]. Biotechnol Bull, 2024, 40(2): 130-145. | |
| [42] | Zhang QY, Chen T, Wang X, et al. Genome-wide identification and expression analyses of homeodomain-leucine zipper family genes reveal their involvement in stress response in apple (Malus×domestica) [J]. Hortic Plant J, 2022, 8(3): 261-278. |
| [43] | Shen W, Li H, Teng RM, et al. Genomic and transcriptomic analyses of HD-Zip family transcription factors and their responses to abiotic stress in tea plant (Camellia sinensis) [J]. Genomics, 2019, 111(5): 1142-1151. |
| [44] | Xie LH, Yan TX, Li L, et al. An HD-ZIP-MYB complex regulates glandular secretory trichome initiation in Artemisia annua [J]. New Phytol, 2021, 231(5): 2050-2064. |
| [45] | Romani F, Ribone PA, Capella M, et al. A matter of quantity: Common features in the drought response of transgenic plants overexpressing HD-Zip I transcription factors [J]. Plant Sci, 2016, 251: 139-154. |
| [46] | He G, Liu P, Zhao H, Sun J. The HD-ZIP II transcription factors regulate plant architecture through the auxin pathway [J]. Int J Mol Sci, 2020, 21(9): 3250. |
| [47] | Scarpella E, Boot KJ, Rueb S, et al. The procambium specification gene Oshox1 promotes polar auxin transport capacity and reduces its sensitivity toward inhibition [J]. Plant Physiol, 2002, 130: 1349-60. |
| [48] | Raza A. Eco-physiological and biochemical responses of rapeseed (Brassica napus L.) to abiotic stresses: consequences and mitigation strategies [J]. J Plant Growth Regul, 2021, 40(4): 1368-1388. |
| [49] | Himmelbach A, Hoffmann T, Leube M, et al. Homeodomain protein ATHB6 is a target of the protein phosphatase ABI1 and regulates hormone responses in Arabidopsis [J]. EMBO J, 2002, 21(12): 3029-3038. |
| [50] | Valdés AE, Overnäs E, Johansson H, et al. The homeodomain-leucine zipper (HD-Zip) class I transcription factors ATHB7 and ATHB12 modulate abscisic acid signalling by regulating protein phosphatase 2C and abscisic acid receptor gene activities [J]. Plant Mol Biol, 2012, 80(4/5): 405-418. |
| [51] | Zhang SX, Haider I, Kohlen W, et al. Function of the HD-Zip Ⅰ gene Oshox22 in ABA-mediated drought and salt tolerances in rice [J]. Plant Mol Biol, 2012, 80(6): 571-585. |
| [52] | Dai MQ, Hu YF, Ma Q, et al. Functional analysis of rice HOMEOBOX4 (Oshox4) gene reveals a negative function in gibberellin responses [J]. Plant Mol Biol, 2008, 66(3): 289-301. |
| [1] | LONG Lin-xi, ZENG Yin-ping, WANG Qian, DENG Yu-ping, GE Min-qian, CHEN Yan-zhuo, LI Xin-juan, YANG Jun, ZOU Jian. Identification of Sunflower GH3 Gene Family and Analysis of Their Function in Flower Development [J]. Biotechnology Bulletin, 2026, 42(1): 125-138. |
| [2] | ZENG Ting, ZHANG Lan, LUO Rui. Functional Analysis of the Transcription Factor MpR2R3-MYB17 in Regulating Gemma Development in Marchantia polymorpha L. [J]. Biotechnology Bulletin, 2026, 42(1): 208-217. |
| [3] | HE Qi-chen, YANG Yang, ALIYA Waili, TANG Xin-yue, LI Zhong-xi, CHEN Yong-kun, CHEN Ling-na. Investigation into the Family Characteristics of the Lavender Copper Amine Oxidase Gene and the Role of LaCuAO1 in Bioamine Degradation [J]. Biotechnology Bulletin, 2026, 42(1): 114-124. |
| [4] | LYU Cheng-cong, HENG Meng, CHEN Si-qi, JIN Xue-hua. Cloning and Functional Analysis of ZhGSTF Related to Anthocyanin Transport Zantedeschia hybrida [J]. Biotechnology Bulletin, 2026, 42(1): 161-169. |
| [5] | YANG Juan, FENG Hui, JI Nai-zhe, SUN Li-ping, WANG Yun, ZHANG Jia-nan, ZHAO Shi-wei. Cloning and Functional Analysis of AP2/ERF Transcription Factors RcERF4 and RcRAP2-12 in Rose [J]. Biotechnology Bulletin, 2026, 42(1): 150-160. |
| [6] | ZHANG Chi-hao, LIU Jin-nan, CHAO Yue-hui. Cloning and Functional Analysis of a bZIP Transcription Factor MtbZIP29 from Medicago truncatula [J]. Biotechnology Bulletin, 2026, 42(1): 241-250. |
| [7] | LIU Jia-li, SONG Jing-rong, ZHAO Wen-yu, ZHANG Xin-yuan, ZHAO Zi-yang, CAO Yi-bo, ZHANG Ling-yun. Identification of the R2R3-MYB Gene and Expression Analysis of Flavonoid Regulatory Genes in Blueberry [J]. Biotechnology Bulletin, 2025, 41(9): 124-138. |
| [8] | LI Yu-zhen, LI Meng-dan, ZHANG Wei, PENG Ting. Functional Study of RmEXPB2 Genein Rosa multiflora Based on the Identification of the Expansin Gene Family in Rosa sp. [J]. Biotechnology Bulletin, 2025, 41(9): 182-194. |
| [9] | WANG Bin, LIN Chong, YUAN Xiao, JIANG Yuan-yuan, WANG Yu-kun, XIAO Yan-hui. Cloning of bHLH Transcription Factor UNE10 and Its Regulatory Roles in the Biosynthesis of Volatile Compounds in Clove Basil [J]. Biotechnology Bulletin, 2025, 41(9): 207-218. |
| [10] | CHEN Qiang, YU Ying-fei, ZHANG Ying, ZHANG Chong. Regulatory Effect of Methyl Jasmonate on Postharvest Chilling Injury in Oriental Melon ‘Emerald’ [J]. Biotechnology Bulletin, 2025, 41(9): 105-114. |
| [11] | CHENG Ting-ting, LIU Jun, WANG Li-li, LIAN Cong-long, WEI Wen-jun, GUO Hui, WU Yao-lin, YANG Jing-fan, LAN Jin-xu, CHEN Sui-qing. Genome-wide Identification of the Chalcone Isomerase Gene Family in Eucommia ulmoides and Analysis of Their Expression Patterns [J]. Biotechnology Bulletin, 2025, 41(9): 242-255. |
| [12] | XU Xiao-ping, YANG Cheng-long, HE Xing, GUO Wen-jie, WU Jian, FANG Shao-zhong. Cloning of the LoAPS1 and Its Function Analysis during the Process of Dormancy Release in Lilium [J]. Biotechnology Bulletin, 2025, 41(9): 195-206. |
| [13] | ZHANG Chao-chao, HAN Kai-yuan, WANG Tong, CHEN Zhong. Cloning and Functional Analysis of PtoYABBY2 and PtoYABBY12 in Populus tomentosa [J]. Biotechnology Bulletin, 2025, 41(9): 256-264. |
| [14] | SHI Fa-chao, JIANG Yong-hua, LIU Hai-lun, WEN Ying-jie, YAN Qian. Cloning and Functional Analysis of LcTFL1 Gene in Litchi chinensis Sonn. [J]. Biotechnology Bulletin, 2025, 41(9): 159-167. |
| [15] | DONG Xiang-xiang, MIAO Bai-ling, XU He-juan, CHEN Juan-juan, LI Liang-jie, GONG Shou-fu, ZHU Qing-song. Bioinformatics Analysis and Flowering Regulation Function of FveBBX20 Gene in Woodland Strawberry [J]. Biotechnology Bulletin, 2025, 41(9): 115-123. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||