








Biotechnology Bulletin ›› 2025, Vol. 41 ›› Issue (12): 342-350.doi: 10.13560/j.cnki.biotech.bull.1985.2025-0543
Previous Articles Next Articles
YANG Hong1,3( ), ZHANG Chao2, XUE Wen-xuan1, HUANG Wei-yuan1, LIN Chun-hua3(
), ZHANG Chao2, XUE Wen-xuan1, HUANG Wei-yuan1, LIN Chun-hua3( ), WANG Li-feng1(
), WANG Li-feng1( )
)
Received:2025-05-26
Online:2025-12-26
Published:2026-01-06
Contact:
LIN Chun-hua, WANG Li-feng
E-mail:yang_hong0317@126.com;lin3286320@hainanu.edu.com;lfwang@catas.cn
YANG Hong, ZHANG Chao, XUE Wen-xuan, HUANG Wei-yuan, LIN Chun-hua, WANG Li-feng. Functional Study of bZIP Transcription Factor CsFcr3 in Regulating the Sensitivity of Colletotrichum siamense to DMI and QoI Fungicides[J]. Biotechnology Bulletin, 2025, 41(12): 342-350.
引物名称 Primer name | 引物序列 Primer sequence (5′-3′) | 用途 Usage |
|---|---|---|
| UF | ACAACAAGCGTTTAGCGTCTC | 扩增目的基因上臂 |
| UR | AGATGTGGGGCACTGTGGCGTTGGCACGGTCGCGCGATGTTGTCCTGGAAGAG | |
| DF | TATTGCACGGGAATTGCATGCTCTCACCAGCCCCCTCTCTCCCCAGTG | 扩增目的基因下臂 |
| DR | GATGCCTAAGCCTGCAAAC | |
| InF | TCACTGGTGGATTGCCGCTTAG | 转化子验证 |
| InR | TGTGCTGTGGTGGTTGCTGAG | |
| OUF | TGTCTTGTCTTGCCTGCAAC | 转化子验证 |
| OUR | AAAGAACTCGGCAGTAAGCTTG | |
| S2F | GGCGGTGCTATCCTTCCCGTGTT | 扩增氯嘧磺隆抗性基因 |
| S2R | GTGAGAGCATGCAATTCCCGTGCAATA | |
| qCsFcr3_F | ATGGACTTCTCCCACCAGTA | RT-qPCR |
| qCsFcr3_R | CGGCAGCGACGAGCTCCTGTAG | |
| qCsActin_F | GATTTGGCACCACACCTTCTACA | 内参 |
| qCsActin_R | TCTCTGTTGGACTTGGGGTTGAT |
Table 1 Primers used in this study
引物名称 Primer name | 引物序列 Primer sequence (5′-3′) | 用途 Usage |
|---|---|---|
| UF | ACAACAAGCGTTTAGCGTCTC | 扩增目的基因上臂 |
| UR | AGATGTGGGGCACTGTGGCGTTGGCACGGTCGCGCGATGTTGTCCTGGAAGAG | |
| DF | TATTGCACGGGAATTGCATGCTCTCACCAGCCCCCTCTCTCCCCAGTG | 扩增目的基因下臂 |
| DR | GATGCCTAAGCCTGCAAAC | |
| InF | TCACTGGTGGATTGCCGCTTAG | 转化子验证 |
| InR | TGTGCTGTGGTGGTTGCTGAG | |
| OUF | TGTCTTGTCTTGCCTGCAAC | 转化子验证 |
| OUR | AAAGAACTCGGCAGTAAGCTTG | |
| S2F | GGCGGTGCTATCCTTCCCGTGTT | 扩增氯嘧磺隆抗性基因 |
| S2R | GTGAGAGCATGCAATTCCCGTGCAATA | |
| qCsFcr3_F | ATGGACTTCTCCCACCAGTA | RT-qPCR |
| qCsFcr3_R | CGGCAGCGACGAGCTCCTGTAG | |
| qCsActin_F | GATTTGGCACCACACCTTCTACA | 内参 |
| qCsActin_R | TCTCTGTTGGACTTGGGGTTGAT |
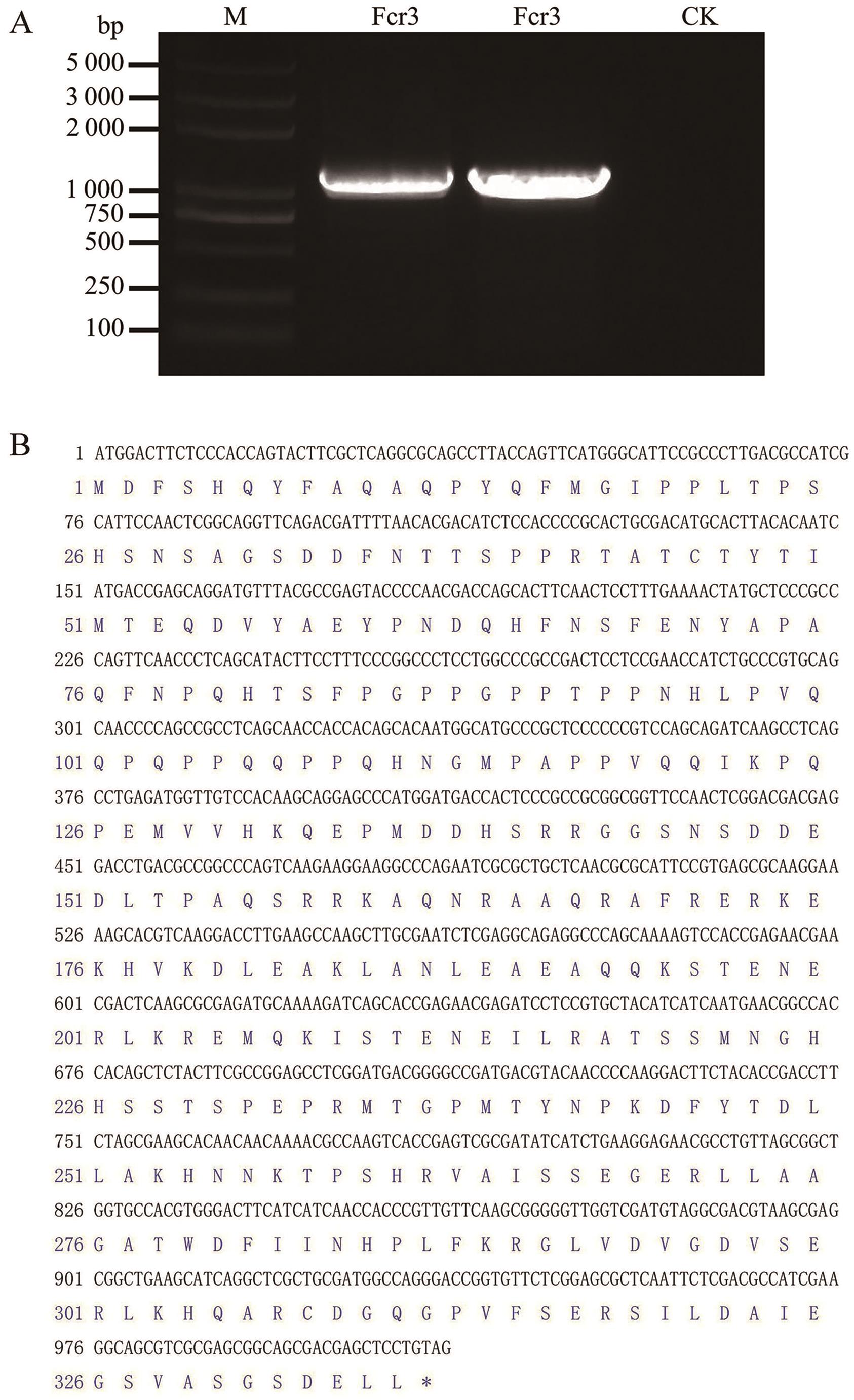
Fig. 1 Full-length cloning of the CsFcr3 geneA: PCR amplification of the CsFcr3 gene. B: Nucleotide sequence and derived amino acids of CsFcr3. M: DL5000 DNA Marker. Fcr3: PCR amplification product of CsFcr3 gene. CK: Negative control
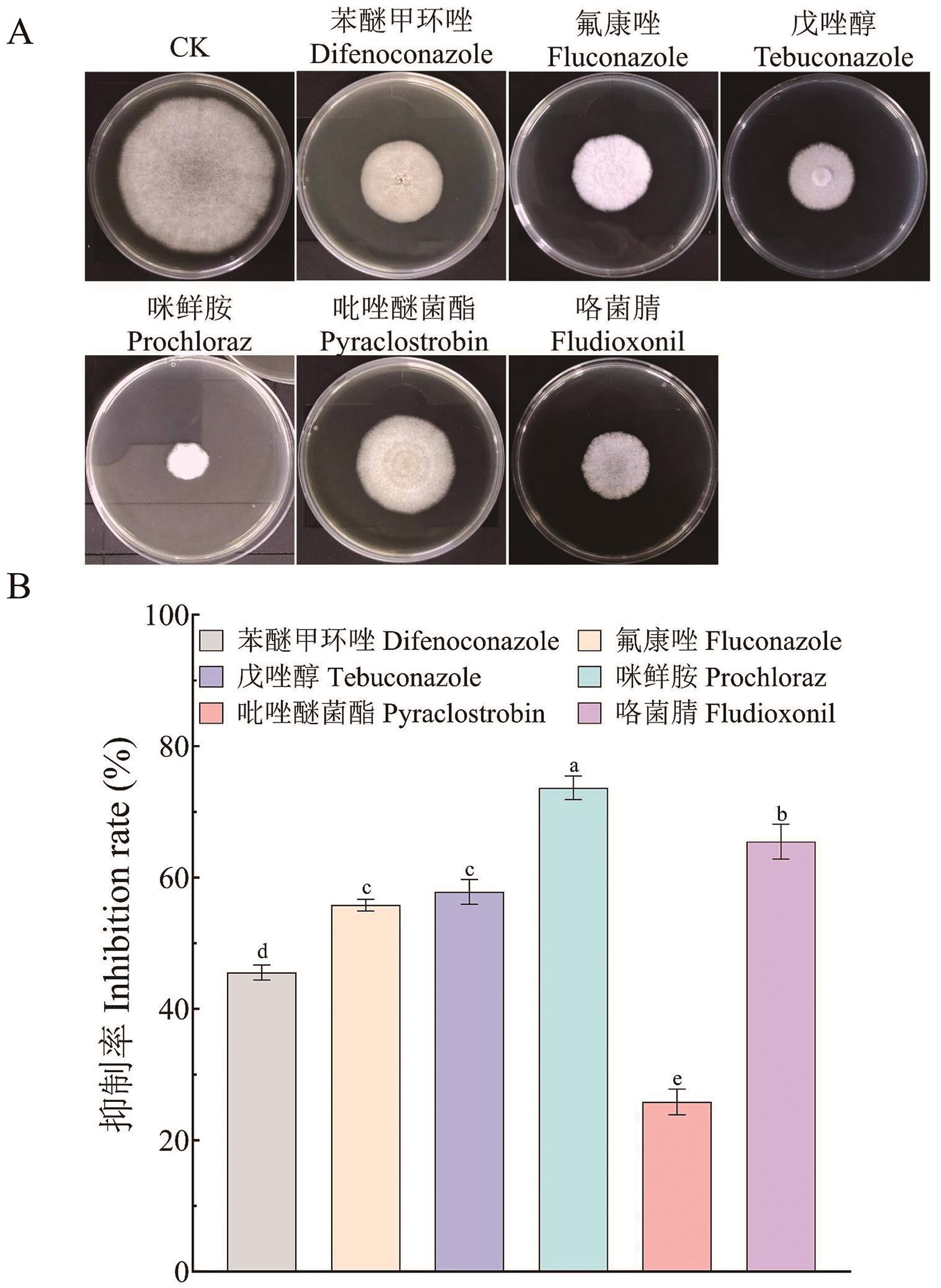
Fig. 3 Phenotype (A) and inhibition rate (B) of HN08 in fungicide treatmentDifferent letters indicate significant differences at P < 0.05, the same below
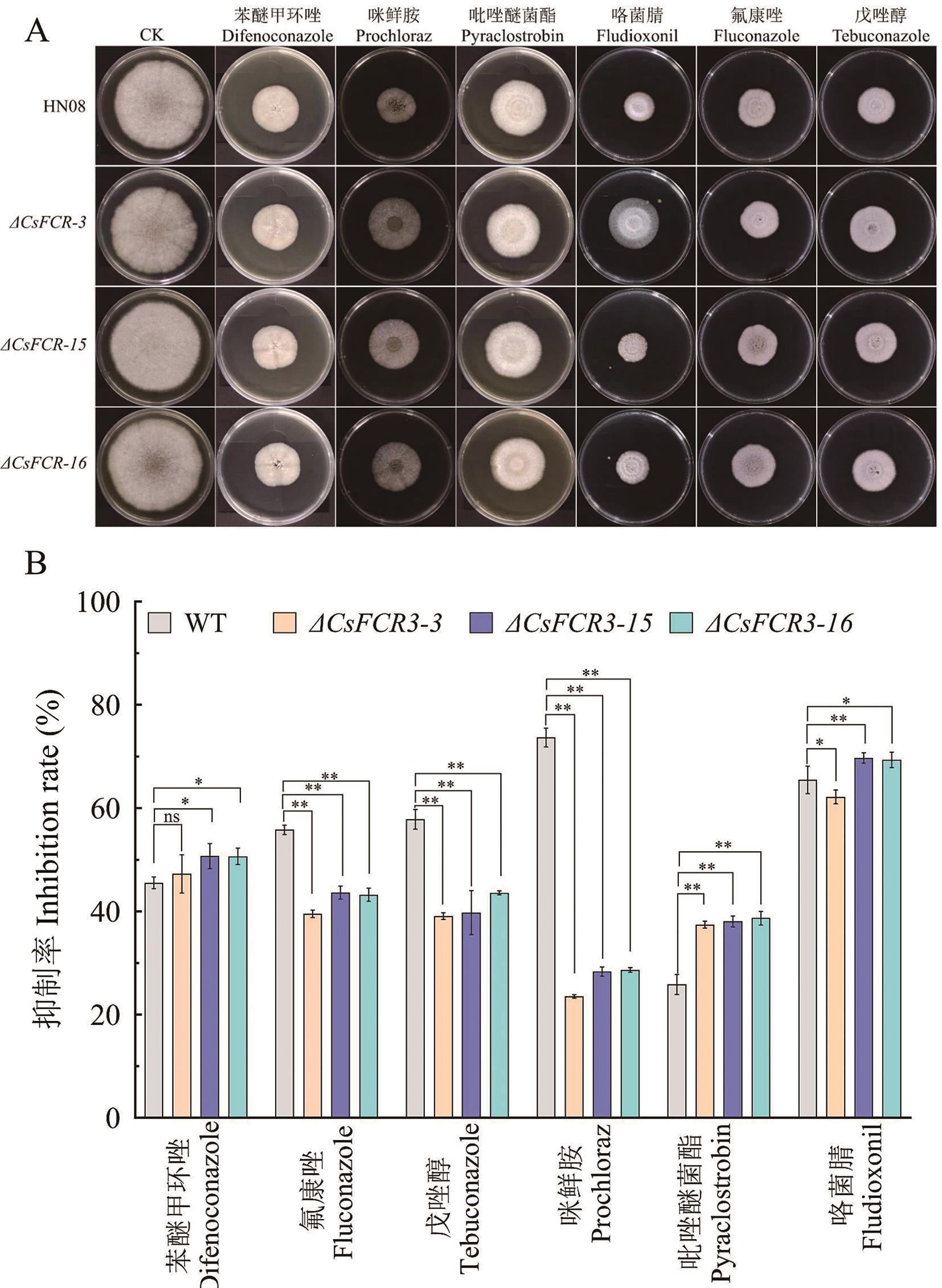
Fig. 6 Sensitivity analysis of ΔCsFcr3s to 6 fungicidesA: Phenotype of ΔCsFcr3s under fungicide stress. B: Inhibition rate of ΔCsFcr3s under fungicide stress, *P < 0.05, **P < 0.01
| [1] | Talhinhas P. Hosts of Colletotrichum [J]. Mycosphere, 2023, 14(si2): 158-261. |
| [2] | 李博勋, 刘先宝, 陈丽琼, 等. 橡胶树主要病害研究现状与展望 [J]. 中国科学: 生命科学, 2024, 54(10): 1798-1813. |
| Li BX, Liu XB, Chen LQ, et al. Current status and prospects of research on main diseases of rubber trees [J]. Sci Sin Vitae, 2024, 54(10): 1798-1813. | |
| [3] | 林春花, 张宇, 刘文波, 等. 我国巴西橡胶树炭疽病的研究进展 [J]. 热带生物学报, 2021, 12(3): 393-402, 268. |
| Lin CH, Zhang Y, Liu WB, et al. Research advances on Colletotrichum leaf fall disease of rubber trees in China [J]. J Trop Biol, 2021, 12(3): 393-402, 268. | |
| [4] | Liu XB, Li BX, Yang Y, et al. Pathogenic adaptations revealed by comparative genome analyses of two Colletotrichum spp., the causal agent of anthracnose in rubber tree [J]. Front Microbiol, 2020, 11: 1484. |
| [5] | Liu XB, Li BX, Cai JM, et al. Colletotrichum species causing anthracnose of rubber trees in China [J]. Sci Rep, 2018, 8: 10435. |
| [6] | Cao XR, Xu XM, Che HY, et al. Three Colletotrichum species, including a new species, are associated to leaf anthracnose of rubber tree in Hainan, China [J]. Plant Dis, 2019, 103(1): 117-124. |
| [7] | 贺春萍, 樊兰艳, 吴贺, 等. 枯草芽孢杆菌Czk1脂肽物质对橡胶树炭疽病和白粉病的抑制效果研究 [J]. 中国农业科技导报, 2023, 25(6): 126-134. |
| He CP, Fan LY, Wu H, et al. Study on inhibitory effect of lipopeptides produced by Bacillus subtilis Czk1 on anthracnose and powdery mildew of rubber tree [J]. J Agric Sci Technol, 2023, 25(6): 126-134. | |
| [8] | 刘凤华, 马迪成, 张晓敏, 等. 植物病原真菌对甾醇脱甲基抑制剂类杀菌剂抗性分子机制研究进展 [J]. 农药学学报, 2022, 24(3): 452-464. |
| Liu FH, Ma DC, Zhang XM, et al. Research progress on molecular resistance mechanism of plant pathogenic fungi to sterol demethylase inhibitors [J]. Chin J Pestic Sci, 2022, 24(3): 452-464. | |
| [9] | 叶滔, 马志强, 毕秋艳, 等. 植物病原真菌对甾醇生物合成抑制剂类(SBIs)杀菌剂的抗药性研究进展 [J]. 农药学学报, 2012, 14(1): 1-16. |
| Ye T, Ma ZQ, Bi QY, et al. Research advances on the resistance of plant pathogenic fungi to SBIs fungicides [J]. Chin J Pestic Sci, 2012, 14(1): 1-16. | |
| [10] | Han YC, Zeng XG, Guo C, et al. Reproduction response of Colletotrichum fungi under the fungicide stress reveals new aspects of chemical control of fungal diseases [J]. Microb Biotechnol, 2022, 15(2): 431-441. |
| [11] | Egner U, Gerbling KP, Hoyer GA, et al. Design of inhibitors of photosystem II using a model of the D1 protein [J]. Pestic Sci, 1996, 47(2): 145-158. |
| [12] | Furukawa K, Randhawa A, Kaur H, et al. Fungal fludioxonil sensitivity is diminished by a constitutively active form of the group III histidine kinase [J]. FEBS Lett, 2012, 586(16): 2417-2422. |
| [13] | 曹学仁, 车海彦, 杨毅, 等. 2014年海南省橡胶炭疽病菌对多菌灵和咪鲜胺的敏感性测定 [J]. 植物病理学报, 2015, 45(6): 626-631. |
| Cao XR, Che HY, Yang Y, et al. Sensitivity of Colletotrichum spp. from Hevea brasiliensis to carbendazim and prochloraz in Hainan Province in China in 2014 [J]. Acta Phytopathol Sin, 2015, 45(6): 626-631. | |
| [14] | 蔡志英, 李加智, 王进强, 等. 橡胶胶孢炭疽菌和尖孢炭疽菌对杀菌剂的敏感性测定 [J]. 云南农业大学学报, 2008, 23(6): 787-790. |
| Cai ZY, Li JZ, Wang JQ, et al. Sensitivity test of Colletotrichum gloeosporioides and Colletotrichum acutatum isolated from rubber to the fungicides [J]. J Yunnan Agric Univ, 2008, 23(6): 787-790. | |
| [15] | 林春花, 徐轩, 戈玉琪, 等. 中国2种橡胶树炭疽病菌对咪鲜胺敏感性比较 [J]. 热带作物学报, 2017, 38(1): 111-115. |
| Lin CH, Xu X, Ge YQ, et al. Comparison of sensitivity between two Colletotrichum species from Hevea brasiliensis to prochloraz in China [J]. Chin J Trop Crops, 2017, 38(1): 111-115. | |
| [16] | Shelest E. Transcription factors in fungi [J]. FEMS Microbiol Lett, 2008, 286(2): 145-151. |
| [17] | Shelest E. Transcription factors in fungi: TFome dynamics, three major families, and dual-specificity TFs [J]. Front Genet, 2017, 8: 53. |
| [18] | Leiter É, Emri T, Pákozdi K, et al. The impact of bZIP Atf1 ortholog global regulators in fungi [J]. Appl Microbiol Biotechnol, 2021, 105(14): 5769-5783. |
| [19] | Takeda T, Toda T, Kominami K, et al. Schizosaccharomyces pombe atf1+ encodes a transcription factor required for sexual development and entry into stationary phase [J]. EMBO J, 1995, 14(24): 6193-6208. |
| [20] | Li YF, Li YS, Lu HH, et al. The bZIP transcription factor ATF1 regulates blue light and oxidative stress responses in Trichoderma guizhouense [J]. mLife, 2023, 2(4): 365-377. |
| [21] | Tang C, Li TY, Klosterman SJ, et al. The bZIP transcription factor VdAtf1 regulates virulence by mediating nitrogen metabolism in Verticillium dahliae [J]. New Phytol, 2020, 226(5): 1461-1479. |
| [22] | Song M, Fang SQ, Li ZG, et al. CsAtf1, a bZIP transcription factor, is involved in fludioxonil sensitivity and virulence in the rubber tree anthracnose fungus Colletotrichum siamense [J]. Fungal Genet Biol, 2022, 158: 103649. |
| [23] | Tang W, Ru YY, Hong L, et al. System-wide characterization of bZIP transcription factor proteins involved in infection-related morphogenesis of Magnaporthe oryzae [J]. Environ Microbiol, 2015, 17(4): 1377-1396. |
| [24] | Hussain S, Tai BW, Hussain A, et al. Genome-wide identification and expression analysis of the basic leucine zipper (bZIP) transcription factor gene family in Fusarium graminearum [J]. Genes, 2022, 13(4): 607. |
| [25] | Yin WB, Reinke AW, Szilágyi M, et al. bZIP transcription factors affecting secondary metabolism, sexual development and stress responses in Aspergillus nidulans [J]. Microbiology, 2013, 159(Pt 1): 77-88. |
| [26] | Zuriegat Q, Zheng YR, Liu H, et al. Current progress on pathogenicity-related transcription factors in Fusarium oxysporum [J]. Mol Plant Pathol, 2021, 22(7): 882-895. |
| [27] | Fernandes L, Rodrigues-Pousada C, Struhl K. Yap, a novel family of eight bZIP proteins in Saccharomyces cerevisiae with distinct biological functions [J]. Mol Cell Biol, 1997, 17(12): 6982-6993. |
| [28] | Chen KH, Miyazaki T, Tsai HF, et al. The bZip transcription factor Cgap1p is involved in multidrug resistance and required for activation of multidrug transporter gene CgFLR1 in Candida glabrata. [J]. Gene, 2007, 386(1/2): 63-72. |
| [29] | Yang XS, Talibi D, Weber S, et al. Functional isolation of the Candida albicans FCR3 gene encoding a bZip transcription factor homologous to Saccharomyces cerevisiae Yap3p [J]. Yeast, 2001, 18(13): 1217-1225. |
| [30] | Yang H, Huang WY, Fan SL, et al. Systematic characterization of the bZIP gene family in Colletotrichum siamense and functional analysis of three family members [J]. Int J Biol Macromol, 2025, 286: 138463. |
| [31] | Guan XL, Song M, Lu JW, et al. The transcription factor CsAtf1 negatively regulates the cytochrome P450 gene CsCyp51G1 to increase fludioxonil sensitivity in Colletotrichum siamense [J]. JoF, 2022, 8(10): 1032. |
| [32] | Zhao QQ, Pei H, Zhou XL, et al. Systematic characterization of bZIP transcription factors required for development and aflatoxin generation by high-throughput gene knockout in Aspergillus flavus [J]. J Fungi, 2022, 8(4): 356. |
| [33] | 张一宾. 甲氧基丙烯酸酯类杀菌剂的全球市场概况及进展 [J]. 世界农药, 2016, 38(4): 30-34. |
| Zhang YB. Development and global market of strobilurin fungicides [J]. World Pestic, 2016, 38(4): 30-34. | |
| [34] | 赵健钦, 金京, 陈杰. QoI类杀菌剂应用与抗性机制研究进展 [J]. 世界农药, 2023, 45(7): 19-30. |
| Zhao JQ, Jin J, Chen J. Research progress on application and resistance of QoI fungicides [J]. World Pestic, 2023, 45(7): 19-30. |
| [1] | ZHANG Ji-chang, XU Yun-feng, JIANG Ling-yan. Optimization of Fermentation Conditions of Endophytic Bacterium ZW21 Isolated from Stylosanthes and Stability Analysis of Antimicrobial Substances [J]. Biotechnology Bulletin, 2025, 41(5): 280-289. |
| [2] | NIU Ruo-yu, GAO Zhan, XIONG Xian-peng, ZHU De, LUO Hao-tian, MA Xue-yuan, HU Guan-jing. Breeding Applications and Prospects of Wild Cotton Germplasm Resources [J]. Biotechnology Bulletin, 2025, 41(4): 21-32. |
| [3] | LIU Qian, MA Lian-jie, ZHANG Hui, WANG Dong, FAN Mao, LIAO Dun-xiu, ZHAO Zheng-wu, LU Wen-cai. Screening, Identification and Control Effects of Biocontrol Strain TN2 against Pepper Anthracnose [J]. Biotechnology Bulletin, 2025, 41(1): 287-297. |
| [4] | XU Wei-fang, LI He-yu, ZHANG Hui, HE Zi-ang, GAO Wen-heng, XIE Zi-yang, WANG Chuan-wen, YIN Deng-ke. Efficacy and Its Mechanism of Bacterial Strain HX0037 on the Control of Anthracnose Disease of Trichosanthes kirilowii Maxim [J]. Biotechnology Bulletin, 2024, 40(4): 228-241. |
| [5] | YANG Yan, HU Yang, LIU Ni-ru, YIN Lu, YANG Rui, WANG Peng-fei, MU Xiao-peng, ZHANG Shuai, CHENG Chun-zhen, ZHANG Jian-cheng. Cloning and Functional Analysis of MbbZIP43 Gene in ‘Hongmantang’ Red-flesh Apple [J]. Biotechnology Bulletin, 2024, 40(2): 146-159. |
| [6] | WU Shuang, LU Rui-lin, FENG Cheng-tian, YUAN Kun, WANG Zhen-hui, LIU Jin-ping, LIU Hui. Expression of HbTRXh5 Gene of Hevea brasiliensis in Yeast and Analysis on Its Resistance to Stress [J]. Biotechnology Bulletin, 2024, 40(12): 136-144. |
| [7] | LI Bai-xue, LI Jin-ling, CHEN Chun-lin, DU Qing-jie, LI Meng, WANG Ji-qing, MA Yong-bin, XIAO Huai-juan. Cloning and Expression Analysis of CabZIP42 Gene in Pepper [J]. Biotechnology Bulletin, 2024, 40(12): 93-101. |
| [8] | ZHANG Le-le, WANG Guan, LIU Feng, HU Han-qiao, REN Lei. Isolation, Identification and Biocontrol Mechanism of an Antagonistic Bacterium Against Anthracnose on Mango Caused by Colletotrichum gloeosporioides [J]. Biotechnology Bulletin, 2023, 39(4): 277-287. |
| [9] | XU Xiao-wen, LI Jin-cang, HAI Du, ZHA Yu-ping, SONG Fei, WANG Yi-xun. Identification and Diversity Analysis of Mycoviruses from the Phytopathogenic Fungus Colletotrichum spp. of Walnut [J]. Biotechnology Bulletin, 2023, 39(3): 278-289. |
| [10] | ZHOU Jia-yan, ZOU Jian, CHEN Wei-ying, WU Yi-chao, CHEN Xi-tong, WANG Qian, ZENG Wen-jing, HU Nan, YANG Jun. Construction of Multi-gene Interference System for Plant and Analysis of Its Application Efficiency [J]. Biotechnology Bulletin, 2023, 39(1): 115-126. |
| [11] | SHI Ya-qian, SHEN Ya-ru, CHEN Man-ying, HE Shu-min, LIU Yu-han, HE Tian-nan, CHEN Qing-xi, WEN Zhi-feng. Molecular Cloning and Expression Analysis of a F-box Protein Gene FnFBOX1 and Its Promoter from Fragaria nilgerrensis [J]. Biotechnology Bulletin, 2022, 38(2): 44-56. |
| [12] | FANG Yuan, WU Xun, LIN Yu, WANG Hai-yan, WU Hui-ping, JU Yu-liang. Duplex-RPA Detection for Bursaphelenchus xylophilus and Bursaphelenchus mucronatus [J]. Biotechnology Bulletin, 2021, 37(7): 183-190. |
| [13] | LI Feng, CHEN Wen-wen, DENG Jiang-li, MAO Qing-li, QUAN Wen-li, MAO Ya-hui. Inhibitory Effect of Resveratrol on Walnut Bacterial Black Spot Pathogen [J]. Biotechnology Bulletin, 2021, 37(6): 58-65. |
| [14] | LIN Jia-ming, GE Hui, LIN Ke-bing, YANG Zhang-wu, ZHOU Chen, WU Jian-shao, WANG Guo-dong, ZHANG Zhe, YANG Qiu-hua, WANG Yi-lei. Isolation,Identification and Antibiotic Sensitivity Analysis of Bacterial Pathogen from Litopenaeus vannamei with Black Gill Disease [J]. Biotechnology Bulletin, 2020, 36(8): 120-128. |
| [15] | FENG Yan-mei, FAN Xing-hui, ZHAN Hui, TENG Shi-yu, YANG Fang, CHEN Shao-hua. Research Progress on Ecotoxicity and Microbial Degradation of Strobilurin Fungicides [J]. Biotechnology Bulletin, 2017, 33(10): 52-58. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||