








Biotechnology Bulletin ›› 2026, Vol. 42 ›› Issue (1): 125-138.doi: 10.13560/j.cnki.biotech.bull.1985.2025-0560
Previous Articles Next Articles
LONG Lin-xi( ), ZENG Yin-ping, WANG Qian, DENG Yu-ping, GE Min-qian, CHEN Yan-zhuo, LI Xin-juan, YANG Jun(
), ZENG Yin-ping, WANG Qian, DENG Yu-ping, GE Min-qian, CHEN Yan-zhuo, LI Xin-juan, YANG Jun( ), ZOU Jian(
), ZOU Jian( )
)
Received:2025-05-31
Online:2026-01-26
Published:2026-02-04
Contact:
YANG Jun, ZOU Jian
E-mail:longlinxi0212@163.com;yangjun@cwnu.edu.cn;zoujian@cwnu.edu.cn
LONG Lin-xi, ZENG Yin-ping, WANG Qian, DENG Yu-ping, GE Min-qian, CHEN Yan-zhuo, LI Xin-juan, YANG Jun, ZOU Jian. Identification of Sunflower GH3 Gene Family and Analysis of Their Function in Flower Development[J]. Biotechnology Bulletin, 2026, 42(1): 125-138.
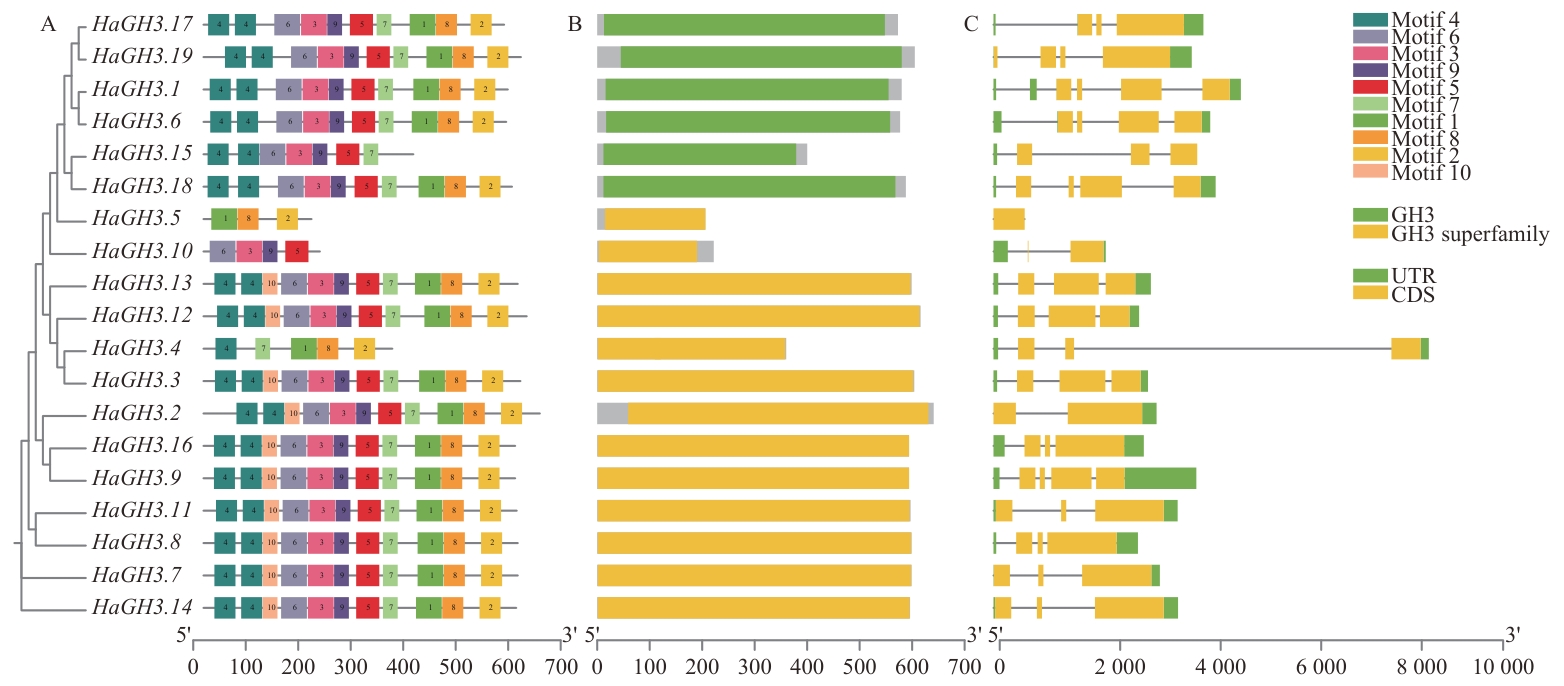
Fig. 2 Gene structure and protein conserved motif distribution of HaGH3A: Conserved motif of protein encoded by HaGH3 gene. B: Conserved domain diagram. C: Gene structure map
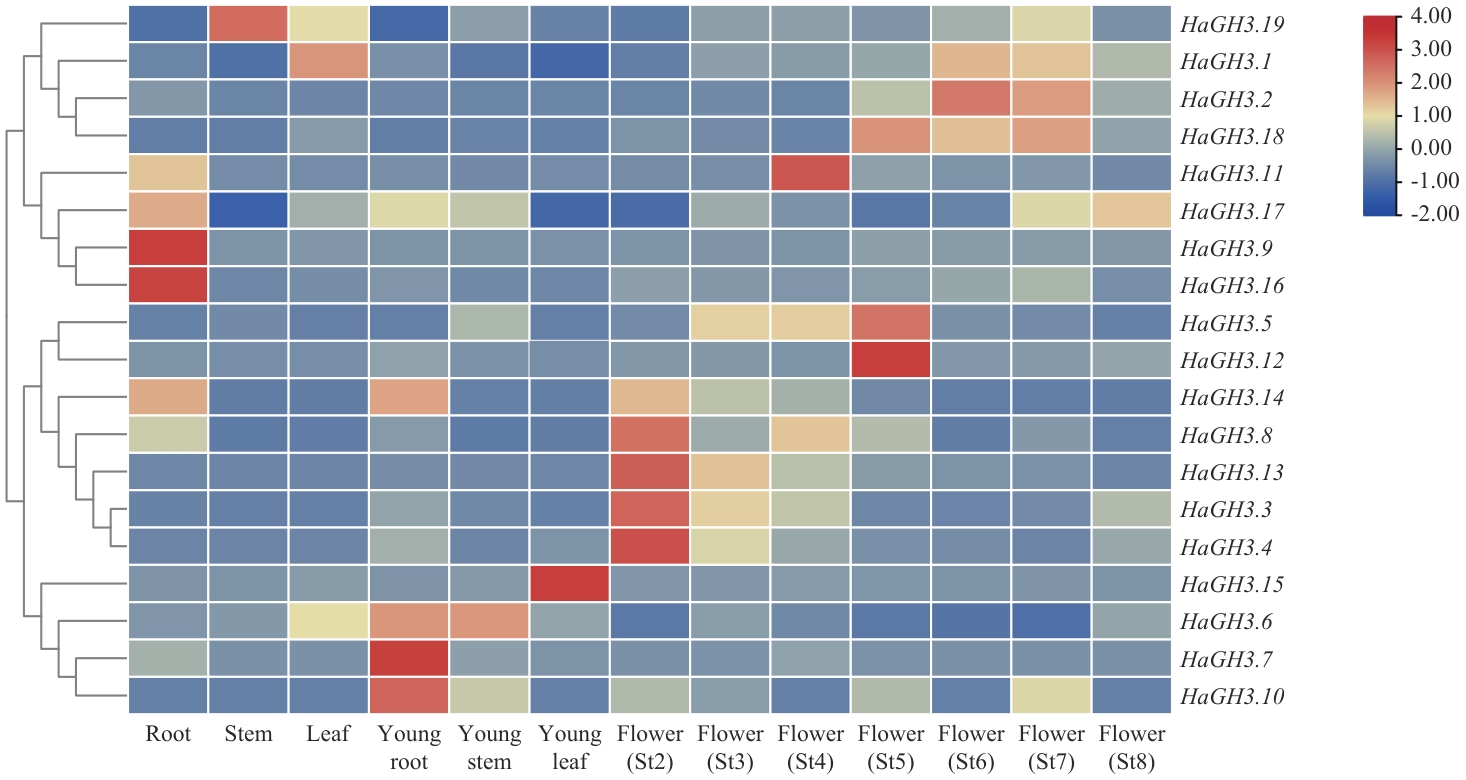
Fig. 3 Expression analysis of HaGH3 family genes in different tissues of WT sunflowerSt2: Initiation phase of flower primordium. St3: Phase of flower organs starting formation. St4: Meiosis phase. St5: Phase of pollen maturity. St6: Floret opening phase. St7: Fruit filling phase. St8: Seed-full phase. The same below
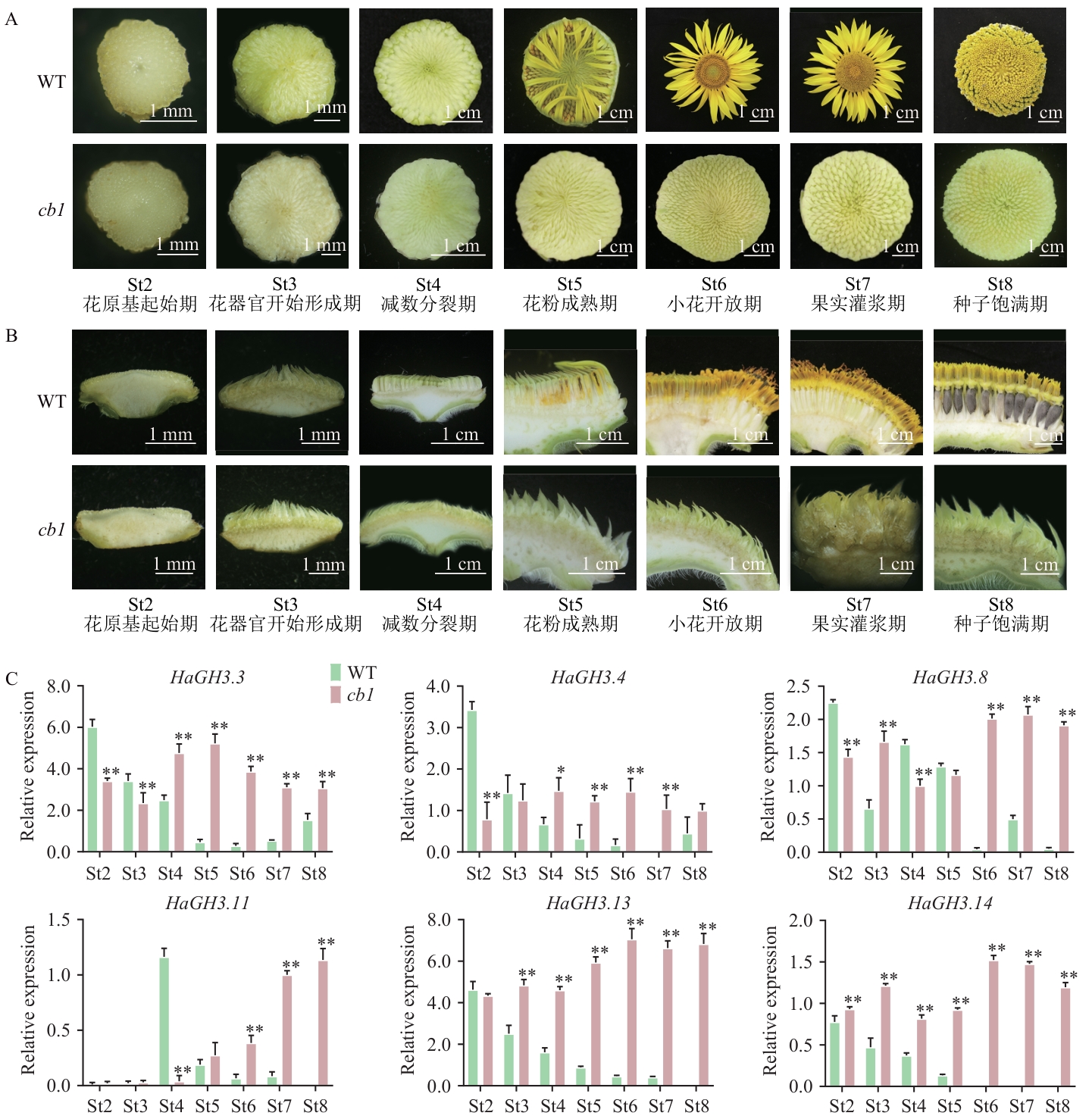
Fig. 4 Differential expression analysis of HaGH3 gene in WT sunflower and cb1 mutantA: Flower disc of WT sunflower and cb1 mutant. B: Longitudinal section of WT sunflower and cb1 mutant inflorescences. C: Relative expression of HaGH3 gene during inflorescence development of WT and cb1 mutants (St2‒St8). All data are mean ±SD (n=3). * P < 0.05, ** P < 0.01. The same below
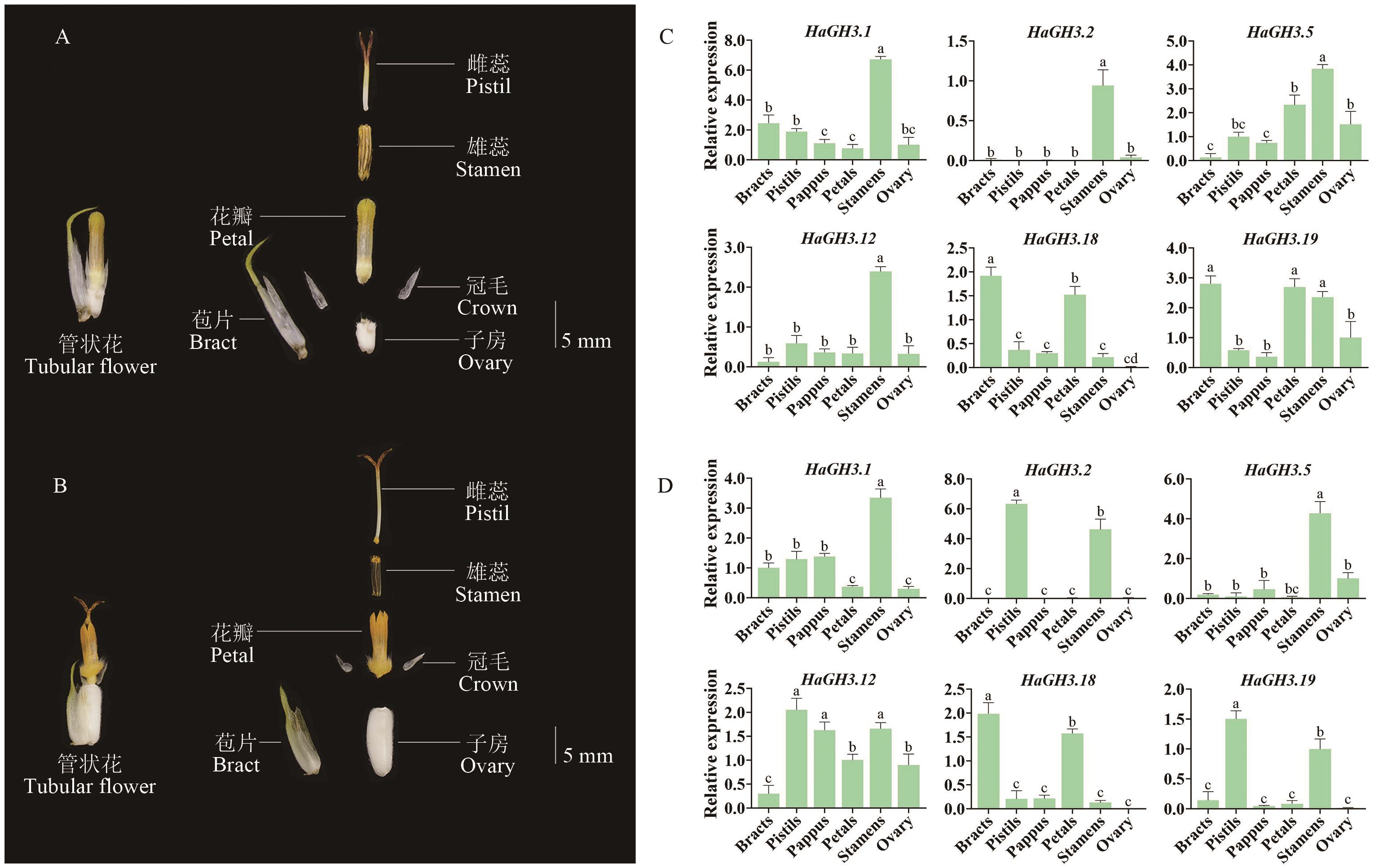
Fig. 5 Expressions of the HaGH3 family in flower organs at different phasesA: Anatomy of tubular flowers at St5 phase of sunflower. B: Anatomy of tubular flowers from the St6 phase of sunflower. C Expressions of sunflower GH3 gene family in flower organs at St5 phase. D: Expressions in flower organs at St6 phase. All data are mean ± SD (n=3). Groups marked with different lowercase letters indicate statistically significant differences (P < 0.05)
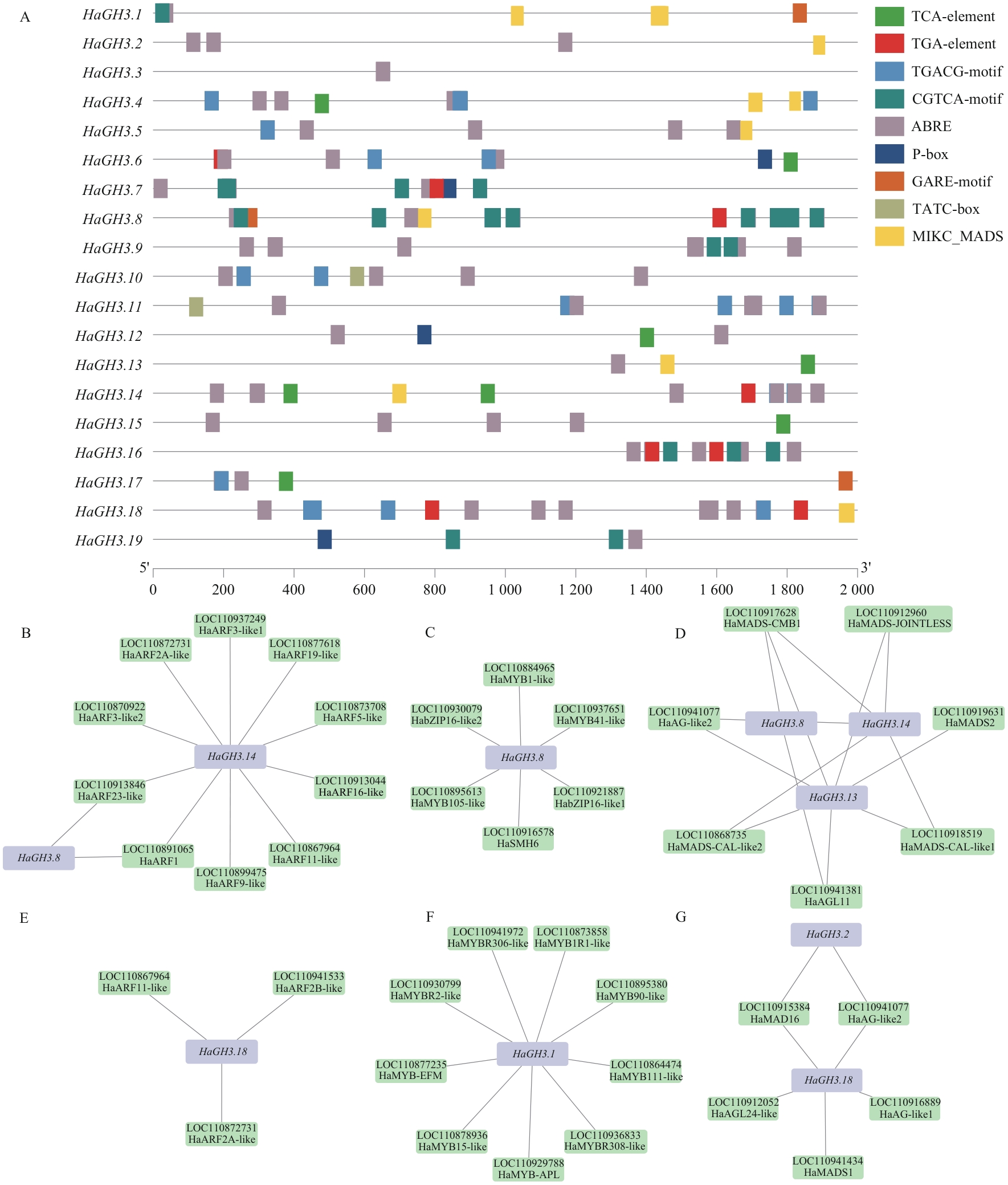
Fig. 6 Prediction analysis of cis-acting elements and regulatory networks of the promoter region of HaGH3 geneA: Cis-acting element of the HaGH3 gene. B: Prediction of interaction between HaGH3.8 and HaGH3.14 with ARF transcription factors. C: Prediction of interaction between HaGH3.8 and MYB and bZIP transcription factors. D: Prediction of interaction between HaGH3.8, HaGH3.13 and HaGH3.14 genes with MIKC_MADS transcription factors. E: Prediction of interaction between HaGH3.18 and ARF transcription factors. F: Prediction of interaction between HaGH3.1 gene and MYB and bZIP transcription factors. G: Prediction of interaction between HaGH3.18 gene and MIKC_MADS transcription factors
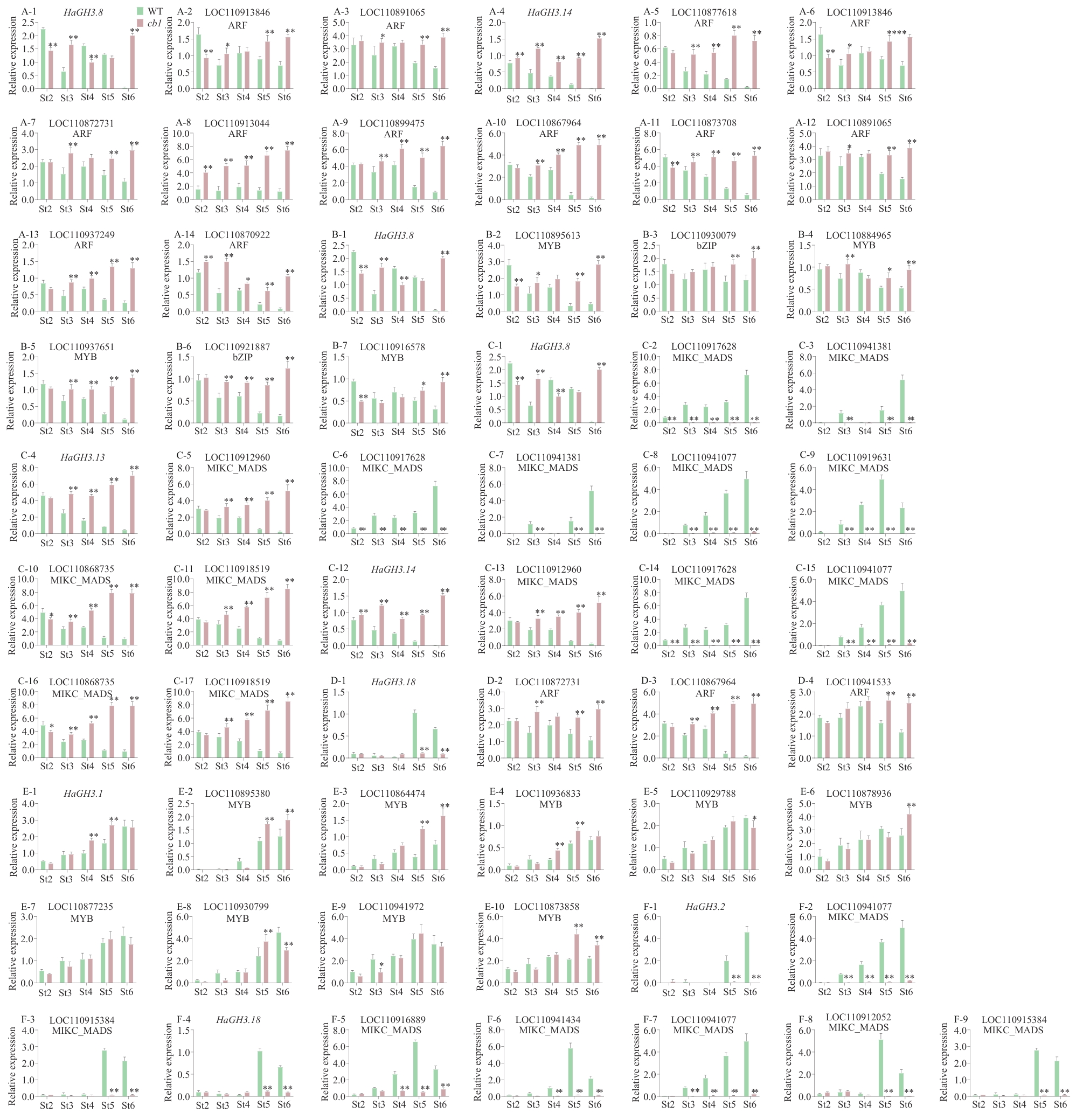
Fig. 7 Relative expressions of HaGH3 gene and its candidate transcription factors involved in the regulation of flower developmentA: Relative expressions of HaGH3.8 and HaGH3.14 and their potential upstream transcriptional regulators ARF. B: Relative expressions of HaGH3.8 and their potential upstream transcriptional regulators MYB and bZIP. C: Relative expressions of HaGH3.8, HaGH3.13 and HaGH3.14 and their potential upstream transcriptional regulators MIKC_MADS. D: Relative expressions of HaGH3.18 and its potential upstream transcriptional regulators ARF. E: Relative expressions of HaGH3.1 and its potential upstream transcriptional regulators MYB and bZIP. F: Relative expressions of HaGH3.18 and its potential upstream transcriptional regulatorsMIKC_MADS
| [1] | Wellmer F, Riechmann JL. Gene networks controlling the initiation of flower development [J]. Trends Genet, 2010, 26(12): 519-527. |
| [2] | Zhang C, Liu XF, Liu Y, et al. An integrated transcriptome and metabolome analysis reveals the gene network regulating flower development in Pogostemon cablin [J]. Front Plant Sci, 2023, 14: 1201486. |
| [3] | Zong J, Wang L, Zhu L, et al. A rice single cell transcriptomic atlas defines the developmental trajectories of rice floret and inflorescence meristems [J]. New Phytol, 2022, 234(2): 494-512. |
| [4] | Cheng XF, Li GF, Krom N, et al. Genetic regulation of flowering time and inflorescence architecture by MtFDa and MtFTa1 in Medicago truncatula [J]. Plant Physiol, 2021, 185(1): 161-178. |
| [5] | 金洲, 卢山, 江俊浩, 等. 园艺植物花芽分化影响因素及机理研究进展 [J]. 园艺学报, 2023, 50(5): 1151-1164. |
| Jin Z, Lu S, Jiang JH, et al. Research progress on influencing factors and mechanisms of flower bud differentiation in horticultural plants [J]. Acta Hortic Sin, 2023, 50(5): 1151-1164. | |
| [6] | 蒋存钰, 申彦华, 王义, 等. 生长素响应因子ARF研究进展 [J/OL]. 分子植物育种, 2022. . |
| Jiang CY, Shen YH, Wang Y, et al. Research progress of auxin response factor ARF [J/OL]. Mol Plant Breed, 2022. . | |
| [7] | Ma CH, Dang KT, Xie QK, et al. Over-expression of ZmIAA29, an AUX/IAA transcription factor, improved maize flowering time [J]. Agronomy, 2023, 13(8): 2028. |
| [8] | 园园, 恩和巴雅尔, 齐艳华. 植物GH3基因家族生物学功能研究进展 [J]. 植物学报, 2023, 58(5): 770-782. |
| Yuan Y, En H, Qi YH. Research advances in biological functions of GH3 gene family in plants [J]. Chin Bull Bot, 2023, 58(5): 770-782. | |
| [9] | Bao DF, Chang SQ, Li XD, et al. Advances in the study of auxin early response genes: Aux/IAA, GH3, and SAUR [J]. Crop J, 2024, 12(4): 964-978. |
| [10] | Staswick PE, Tiryaki I, Rowe ML. Jasmonate response locus JAR1 and several related Arabidopsis genes encode enzymes of the firefly luciferase superfamily that show activity on jasmonic, salicylic, and indole-3-acetic acids in an assay for adenylation [J]. Plant Cell, 2002, 14(6): 1405-1415. |
| [11] | Hagen G, Guilfoyle T. Auxin-responsive gene expression: genes, promoters and regulatory factors [J]. Plant Mol Biol, 2002, 49(3/4): 373-385. |
| [12] | Jagadeeswaran G, Raina S, Acharya BR, et al. Arabidopsis GH3-LIKE DEFENSE GENE-1 is required for accumulation of salicylic acid, activation of defense responses and resistance to Pseudomonas syringae [J]. Plant J, 2007, 51(2): 234-246. |
| [13] | Nobuta RAO K. The GH3 acyl adenylase family member PBS3 regulates salicylic acid-dependent defense responses in Arabidopsis [J]. Plant Physiol, 2007, 144(2): 1144-1156. |
| [14] | Woodward AW, Bartel B. Auxin: regulation, action, and interaction [J]. Ann Bot, 2005, 95(5): 707-735. |
| [15] | Nakazawa M, Yabe N, Ichikawa T, et al. DFL1 an auxin-responsive GH3 gene homologue, negatively regulates shoot cell elongation and lateral root formation, and positively regulates the light response of hypocotyl length [J]. Plant J, 2001, 25(2): 213-221. |
| [16] | Zhao SQ, Xiang JJ, Xue HW. Studies on the rice LEAF INCLINATION1 (LC1), an IAA-amido synthetase, reveal the effects of auxin in leaf inclination control [J]. Mol Plant, 2013, 6(1): 174-187. |
| [17] | Du H, Wu N, Fu J, et al. A GH3 family member, OsGH3-2, modulates auxin and abscisic acid levels and differentially affects drought and cold tolerance in rice [J]. J Exp Bot, 2012, 63(18): 6467-6480. |
| [18] | Delfin JC, Kanno Y, Seo M, et al. AtGH3.10 is another jasmonic acid-amido synthetase in Arabidopsis thaliana [J]. Plant J, 2022, 110(4): 1082-1096. |
| [19] | Zhang SW, Li CH, Cao J, et al. Altered architecture and enhanced drought tolerance in rice via the down-regulation of indole-3-acetic acid by TLD1/OsGH3.13 activation [J]. Plant Physiol, 2009, 151(4): 1889-1901. |
| [20] | Singh VK, Jain M, Garg R. Genome-wide analysis and expression profiling suggest diverse roles of GH3 genes during development and abiotic stress responses in legumes [J]. Front Plant Sci, 2014, 5: 789. |
| [21] | 周苹, 唐冬英, 郭明, 等. 拟南芥GH3.9基因的过表达及其表型分析 [J]. 西北植物学报, 2015, 35(3): 454-458. |
| Zhou P, Tang DY, Guo M, et al. Overexpression and phenotype analysis of GH3.9 gene in Arabidopsis thaliana [J]. Acta Bot Boreali Occidentalia Sin, 2015, 35(3): 454-458. | |
| [22] | Yadav SR, Khanday I, Majhi BB, et al. Auxin-responsive OsMGH3, a common downstream target of OsMADS1 and OsMADS6, controls rice floret fertility [J]. Plant Cell Physiol, 2011, 52(12): 2123-2135. |
| [23] | Liu SY, Zhang CB, Guo F, et al. A systematical genome-wide analysis and screening of WRKY transcription factor family engaged in abiotic stress response in sweetpotato [J]. BMC Plant Biol, 2022, 22(1): 616. |
| [24] | Guo RP, Hu Y, Aoi Y, et al. Local conjugation of auxin by the GH3 amido synthetases is required for normal development of roots and flowers in Arabidopsis [J]. Biochem Biophys Res Commun, 2022, 589: 16-22. |
| [25] | Riemann M, Riemann M, Takano M. Rice JASMONATE RESISTANT 1 is involved in phytochrome and jasmonate signalling [J]. Plant Cell Environ, 2008, 31(6): 783-792. |
| [26] | Zhang SN, Wang SK, Xu YX, et al. The auxin response factor, OsARF19, controls rice leaf angles through positively regulating OsGH3-5 and OsBRI1 [J]. Plant Cell Environ, 2015, 38(4): 638-654. |
| [27] | 周淑瑶, 李建明, 毛娟. AtGH3.17调控拟南芥生长素和油菜素甾醇的响应 [J]. 植物学报, 2023, 58(3): 373-384. |
| Zhou SY, Li JM, Mao J. AtGH3.17-mediated regulation of auxin and brassinosteroid response in Arabidopsis thaliana [J]. Chin Bull Bot, 2023, 58(3): 373-384. | |
| [28] | Mäkilä R, Wybouw B, Smetana O, et al. Gibberellins promote polar auxin transport to regulate stem cell fate decisions in cambium [J]. Nat Plants, 2023, 9(4): 631-644. |
| [29] | 邹礼平, 潘铖, 王梦馨, 等. 激素调控植物成花机理研究进展 [J]. 遗传, 2020, 42(8): 739-751. |
| Zou LP, Pan C, Wang MX, et al. Progress on the mechanism of hormones regulating plant flower formation [J]. Hereditas, 2020, 42(8): 739-751. | |
| [30] | Alejandra Freire-Rios, Keita Tanaka, Crespo, Isidro, et al. Architecture of DNA elements mediating ARF transcription factor binding and auxin-responsive gene expression in Arabidopsis [J]. Proc Natl Acad Sci U S A, 2020, 117(39): 24557-24566. |
| [31] | 宋松泉, 刘军, 黄荟, 等. 赤霉素代谢与信号转导及其调控种子萌发与休眠的分子机制 [J]. 中国科学: 生命科学, 2020, 50(6): 599-615. |
| Song SQ, Liu J, Huang H, et al. Gibberellin metabolism and signal transduction and its molecular mechanism of regulating seed germination and dormancy [J]. Sci China Ser C, 2020, 50(6): 599-615. | |
| [32] | Hernández-García J, Briones-Moreno A, Blázquez MA. Origin and evolution of gibberellin signaling and metabolism in plants [J]. Semin Cell Dev Biol, 2021, 109: 46-54. |
| [33] | Tsuji H, Aya K, Ueguchi-Tanaka M, et al. GAMYB controls different sets of genes and is differentially regulated by microRNA in aleurone cells and anthers [J]. Plant J, 2006, 47(3): 427-444. |
| [34] | Iven T, Strathmann A, Böttner S, et al. Homo- and heterodimers of tobacco bZIP proteins counteract as positive or negative regulators of transcription during pollen development [J]. Plant J, 2010, 63(1): 155-166. |
| [35] | 王莹, 穆艳霞, 王锦. MADS-box基因家族调控植物花器官发育研究进展 [J]. 浙江农业学报, 2021, 33(6): 1149-1158. |
| Wang Y, Mu YX, Wang J. Research progress of floral development regulation by MADS-box gene family [J]. Acta Agric Zhejiangensis, 2021, 33(6): 1149-1158. |
| [1] | ZENG Ting, ZHANG Lan, LUO Rui. Functional Analysis of the Transcription Factor MpR2R3-MYB17 in Regulating Gemma Development in Marchantia polymorpha L. [J]. Biotechnology Bulletin, 2026, 42(1): 208-217. |
| [2] | ZHANG Chi-hao, LIU Jin-nan, CHAO Yue-hui. Cloning and Functional Analysis of a bZIP Transcription Factor MtbZIP29 from Medicago truncatula [J]. Biotechnology Bulletin, 2026, 42(1): 241-250. |
| [3] | WU Cui-cui, CHEN Deng-ke, LAN Gang, XIA Zhi, LI Peng-bo. Bioinformatics Analysis of Peanut Transcription Factor AhHDZ70 and Its Tolerances to Salt and Drought [J]. Biotechnology Bulletin, 2026, 42(1): 198-207. |
| [4] | CHEN Qiang, YU Ying-fei, ZHANG Ying, ZHANG Chong. Regulatory Effect of Methyl Jasmonate on Postharvest Chilling Injury in Oriental Melon ‘Emerald’ [J]. Biotechnology Bulletin, 2025, 41(9): 105-114. |
| [5] | WANG Bin, LIN Chong, YUAN Xiao, JIANG Yuan-yuan, WANG Yu-kun, XIAO Yan-hui. Cloning of bHLH Transcription Factor UNE10 and Its Regulatory Roles in the Biosynthesis of Volatile Compounds in Clove Basil [J]. Biotechnology Bulletin, 2025, 41(9): 207-218. |
| [6] | LI Kai-jie, WU Yao, LI Dan-dan. Cloning of Gene CtbHLH128 in Safflower and Response Function Regulating Drought Stress [J]. Biotechnology Bulletin, 2025, 41(8): 234-241. |
| [7] | ZENG Dan, HUANG Yuan, WANG Jian, ZHANG Yan, LIU Qing-xia, GU Rong-hui, SUN Qing-wen, CHEN Hong-yu. Genome-wide Identification and Expression Analysis of bZIP Transcription Factor Family in Dendrobium officinale [J]. Biotechnology Bulletin, 2025, 41(8): 197-210. |
| [8] | HUANG Shi-yu, TIAN Shan-shan, YANG Tian-wei, GAO Man-rong, ZHANG Shang-wen. Genome-wide Identification and Expression Pattern Analysis of WRI1 Gene Family in Erythropalum scandens [J]. Biotechnology Bulletin, 2025, 41(8): 242-254. |
| [9] | GAO Jing, CHENG Yi-cun, GAO Ming, ZHAO Yun-xiao, WANG Yang-dong. Regulation of Plant Tannin Synthesis and Mechanisms of Its Responses to Environment [J]. Biotechnology Bulletin, 2025, 41(7): 49-59. |
| [10] | NIU Jing-ping, ZHAO Jing, GUO Qian, WANG Shu-hong, ZHAO Jin-zhong, DU Wei-jun, YIN Cong-cong, YUE Ai-qin. Identification and Induced Expression Analysis of Transcription Factors NAC in Soybean Resistance to Soybean Mosaic Virus Based on WGCNA [J]. Biotechnology Bulletin, 2025, 41(7): 95-105. |
| [11] | LI Xia, ZHANG Ze-wei, LIU Ze-jun, WANG Nan, GUO Jiang-bo, XIN Cui-hua, ZHANG Tong, JIAN Lei. Cloning and Functional Study of Transcription Factor StMYB96 in Potato [J]. Biotechnology Bulletin, 2025, 41(7): 181-192. |
| [12] | GONG Yu-han, CHEN Lan, SHANGFANG Hui-zi, HAO Ling-ying, LIU Shuo-qian. Identification and Expression Profile Analysis of the TRB Gene Family in Tea Plant [J]. Biotechnology Bulletin, 2025, 41(7): 214-225. |
| [13] | WEI Yu-jia, LI Yan, KANG Yu-han, GONG Xiao-nan, DU Min, TU Lan, SHI Peng, YU Zi-han, SUN Yan, ZHANG Kun. Cloning and Expression Analysis of the CrMYB4 Gene in Carex rigescens [J]. Biotechnology Bulletin, 2025, 41(7): 248-260. |
| [14] | LI Rui, HU Ting, CHEN Shu-wei, WANG Yao, WANG Ji-ping. Positive Regulation of Anthocyanin Biosynthesis by PfMYB80 Transcription Factor in Perilla frutescens [J]. Biotechnology Bulletin, 2025, 41(6): 243-255. |
| [15] | GUO Tao, AI Li-jiao, ZOU Shi-hui, ZHOU Ling, LI Xue-mei. Functional Study of CjRAV1 from Camellia japonica in Regulating Flowering Delay [J]. Biotechnology Bulletin, 2025, 41(6): 208-217. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||