








Biotechnology Bulletin ›› 2026, Vol. 42 ›› Issue (1): 241-250.doi: 10.13560/j.cnki.biotech.bull.1985.2025-0482
Previous Articles Next Articles
ZHANG Chi-hao( ), LIU Jin-nan(
), LIU Jin-nan( ), CHAO Yue-hui(
), CHAO Yue-hui( )
)
Received:2025-05-13
Online:2026-01-26
Published:2025-12-23
Contact:
CHAO Yue-hui
E-mail:zzzch0127@126.com;chaoyuehui@bjfu.edu.cn
ZHANG Chi-hao, LIU Jin-nan, CHAO Yue-hui. Cloning and Functional Analysis of a bZIP Transcription Factor MtbZIP29 from Medicago truncatula[J]. Biotechnology Bulletin, 2026, 42(1): 241-250.
引物名称 Primer name | 序列 Sequence(5′-3′) | 用途 Usage |
|---|---|---|
| MtbZIP29-F | GATAAGCGCCTGCTCCTT | 基因克隆 Gene cloning |
| MtbZIP29-R | ACGACCTCCAATCCAACC | |
| M13-47 | CAGGGTTTTCCCAGTCACG | |
| M13-48 | GAGCGGATAACAATTTCACAC | |
| 3302-bZIP29-F | CATGGTAGATCTGACTAGTATGGAAGATACTGAAGCAGCTTCATCTTTCAAAATG | 构建亚细胞定位载体 Construction of subcellular localization vector |
| 3302-bZIP29-R | GGCCTCCTAGGTACACGCGTTTGCTTTAACTCATTCTTTGAGTTGGCATT | |
| BDbZIP29-F | TCAGAGGAGGACCTGCATATGGAAGATACTGAAGCAGCT | 构建酵母表达载体 Construction of yeast expression vector |
| BD-bZIP29-R | CGCTGCAGGTCGACGGATCCCTATTGCTTTAACTCATTCTTTGAGTTG | |
| BD-bZIP29-U-F | GCCATGGAGGCCGAATTCCCGATGGAAGATACTGAAGCAGC | 构建酵母表达载体(N端) Construction of yeast expression vector (N-terminus) |
| BD-ZIP29-U-R | CTGCAGGTCGACGGATCCCCTTACACAACTTCACCTTCCGGCG | |
| BD-bZIP29-D-F | GCCATGGAGGCCGAATTCCCGGATGATCTTTTTTCTGCTTA | 构建酵母表达载体(C端) Construction of yeast expression vector (C-terminus) |
| BD-bZIP29-D-R | CTGCAGGTCGACGGATCCCCCTATTGCTTTAACTCATTCT | |
| MtbZIP29 qRT-F | TGGGAATGGTGAGTTTAG | 基因表达分析 Gene expression analysis |
| MtbZIP29 qRT-R | CAGAAAGTGTAGTGGCTTC | |
| MtActin-F | TGATCTGGCTGGTCGTGACCTTA | 蒺藜苜蓿内参基因表达分析 Expression analysis of Medicago truncatula internal reference gene |
| MtActin-R | ATGCCTGCTGCTTCCATTCCTAT | |
| Nt-bZIP29-F | ATCAGATCCCGGTTTCAC | 转基因烟草PCR检测 PCR analysis of transgenic tobacco |
| Nt-bZIP29-R | CTATCATCAGTAGCCGCGT |
Table 1 Primers and sequences
引物名称 Primer name | 序列 Sequence(5′-3′) | 用途 Usage |
|---|---|---|
| MtbZIP29-F | GATAAGCGCCTGCTCCTT | 基因克隆 Gene cloning |
| MtbZIP29-R | ACGACCTCCAATCCAACC | |
| M13-47 | CAGGGTTTTCCCAGTCACG | |
| M13-48 | GAGCGGATAACAATTTCACAC | |
| 3302-bZIP29-F | CATGGTAGATCTGACTAGTATGGAAGATACTGAAGCAGCTTCATCTTTCAAAATG | 构建亚细胞定位载体 Construction of subcellular localization vector |
| 3302-bZIP29-R | GGCCTCCTAGGTACACGCGTTTGCTTTAACTCATTCTTTGAGTTGGCATT | |
| BDbZIP29-F | TCAGAGGAGGACCTGCATATGGAAGATACTGAAGCAGCT | 构建酵母表达载体 Construction of yeast expression vector |
| BD-bZIP29-R | CGCTGCAGGTCGACGGATCCCTATTGCTTTAACTCATTCTTTGAGTTG | |
| BD-bZIP29-U-F | GCCATGGAGGCCGAATTCCCGATGGAAGATACTGAAGCAGC | 构建酵母表达载体(N端) Construction of yeast expression vector (N-terminus) |
| BD-ZIP29-U-R | CTGCAGGTCGACGGATCCCCTTACACAACTTCACCTTCCGGCG | |
| BD-bZIP29-D-F | GCCATGGAGGCCGAATTCCCGGATGATCTTTTTTCTGCTTA | 构建酵母表达载体(C端) Construction of yeast expression vector (C-terminus) |
| BD-bZIP29-D-R | CTGCAGGTCGACGGATCCCCCTATTGCTTTAACTCATTCT | |
| MtbZIP29 qRT-F | TGGGAATGGTGAGTTTAG | 基因表达分析 Gene expression analysis |
| MtbZIP29 qRT-R | CAGAAAGTGTAGTGGCTTC | |
| MtActin-F | TGATCTGGCTGGTCGTGACCTTA | 蒺藜苜蓿内参基因表达分析 Expression analysis of Medicago truncatula internal reference gene |
| MtActin-R | ATGCCTGCTGCTTCCATTCCTAT | |
| Nt-bZIP29-F | ATCAGATCCCGGTTTCAC | 转基因烟草PCR检测 PCR analysis of transgenic tobacco |
| Nt-bZIP29-R | CTATCATCAGTAGCCGCGT |
| 用途 Usage | 网址 Website |
|---|---|
| 启动子及顺式作用元件预测 Promoter and cis-acting element prediction | PlantCARE, http://bioinformatics.psb.ugent.be/webtools/plantcare/html |
| 系统进化树分析 Phylogenetic tree analysis | NCBI, https://www.ncbi.nlm.nih.gov/ |
| 蛋白质结构域分析 Protein structure domain analysis | UniProt, https://www.uniprot.org/ |
| 蛋白质二级结构预测 Protein secondary structure prediction | SOPMA, https://npsa.lyon.inserm.fr/cgibin/npsa_automat.pl page=/NPSA/npsa_sopma.html |
| 蛋白质三级结构预测 Protein tertiary structure prediction | Swissmodel, https://swissmodel.expasy.org/ |
| 蛋白质理化性质分析 Analysis of physicochemical properties of proteins | Expasy-ProtParam, http://web.expasy.org/protparam Expasy-ProtScale, http://web.expasy.org/protscale |
Table 2 Bioinformation analysis of MtbZIP29
| 用途 Usage | 网址 Website |
|---|---|
| 启动子及顺式作用元件预测 Promoter and cis-acting element prediction | PlantCARE, http://bioinformatics.psb.ugent.be/webtools/plantcare/html |
| 系统进化树分析 Phylogenetic tree analysis | NCBI, https://www.ncbi.nlm.nih.gov/ |
| 蛋白质结构域分析 Protein structure domain analysis | UniProt, https://www.uniprot.org/ |
| 蛋白质二级结构预测 Protein secondary structure prediction | SOPMA, https://npsa.lyon.inserm.fr/cgibin/npsa_automat.pl page=/NPSA/npsa_sopma.html |
| 蛋白质三级结构预测 Protein tertiary structure prediction | Swissmodel, https://swissmodel.expasy.org/ |
| 蛋白质理化性质分析 Analysis of physicochemical properties of proteins | Expasy-ProtParam, http://web.expasy.org/protparam Expasy-ProtScale, http://web.expasy.org/protscale |
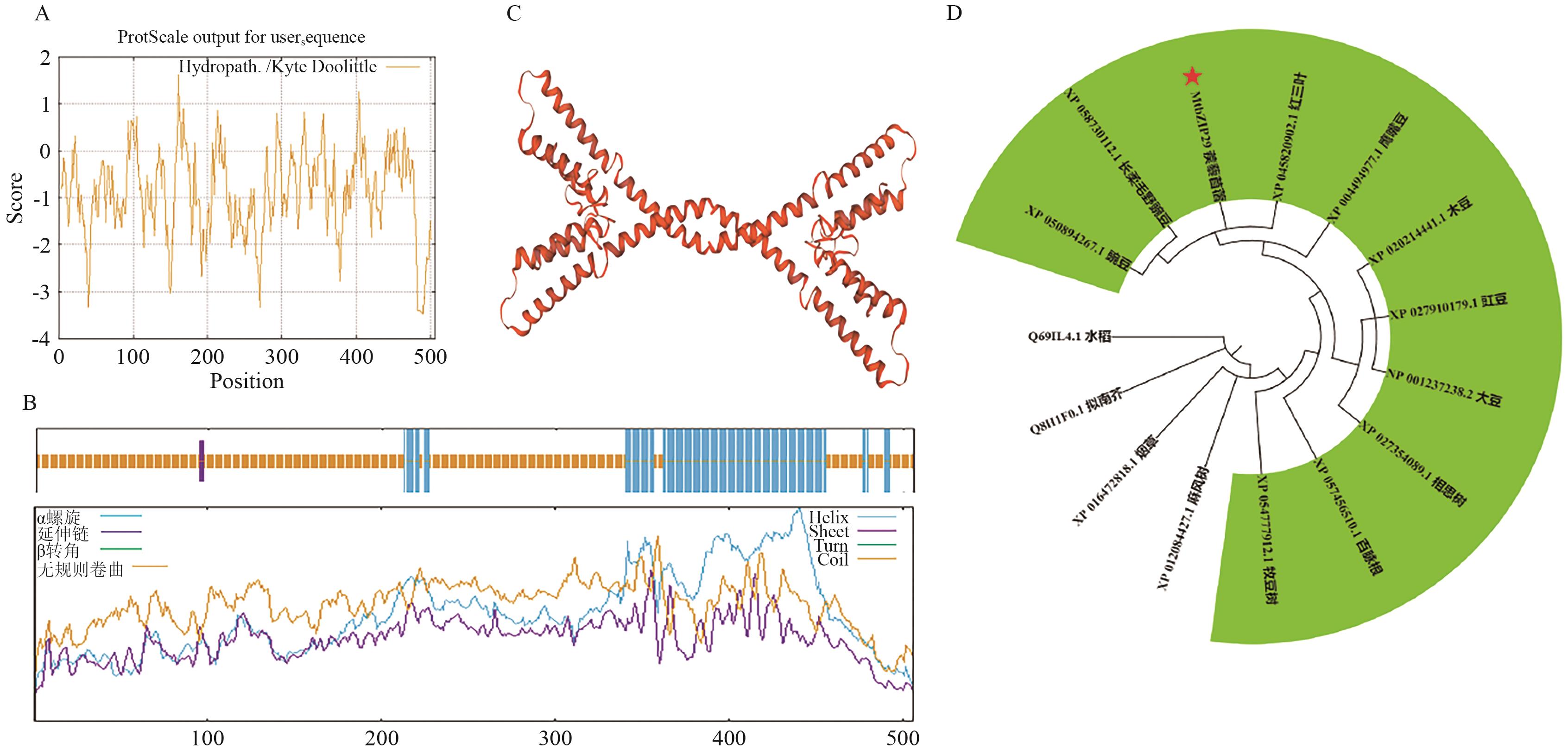
Fig. 3 Protein structure and genetic evolutionary analysis of protein MtbZIP29A: Hydrophobicity analysis of protein. B: The secondary structure of protein. C: The tertiary structure of protein. D: Phylogenetic tree analysis
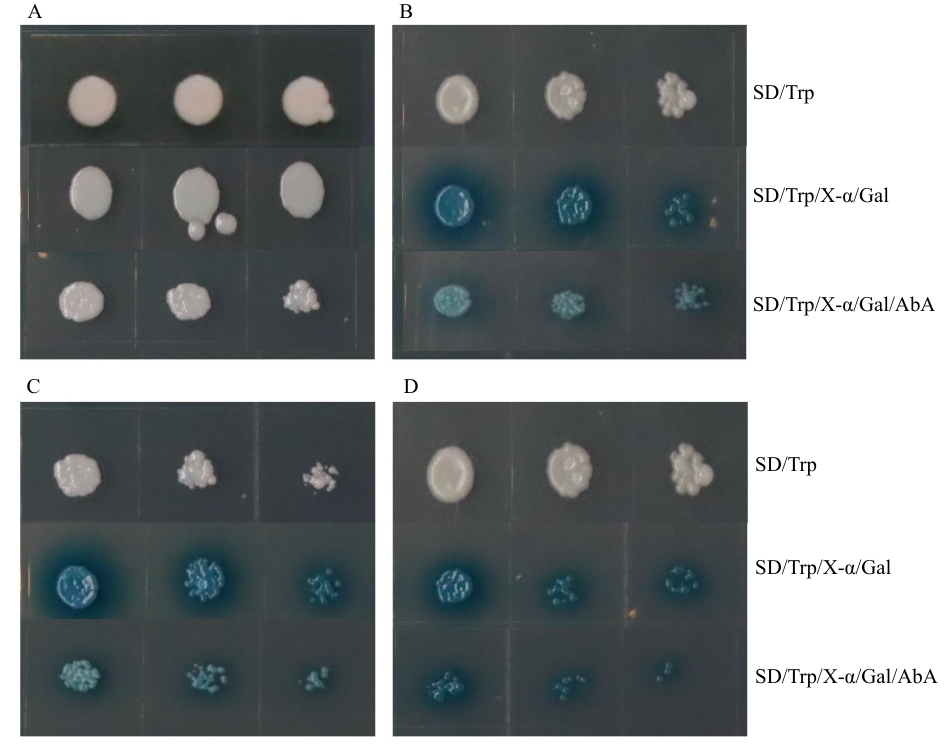
Fig. 6 MtbZIP29 self-activation assayA: Yeast transformed with BD empty vector. B: Yeast transformed with BD-bZIP29. C: Yeast transformed with BD-bZIP29-N. D: Yeast transformed with BD-bZIP29-C
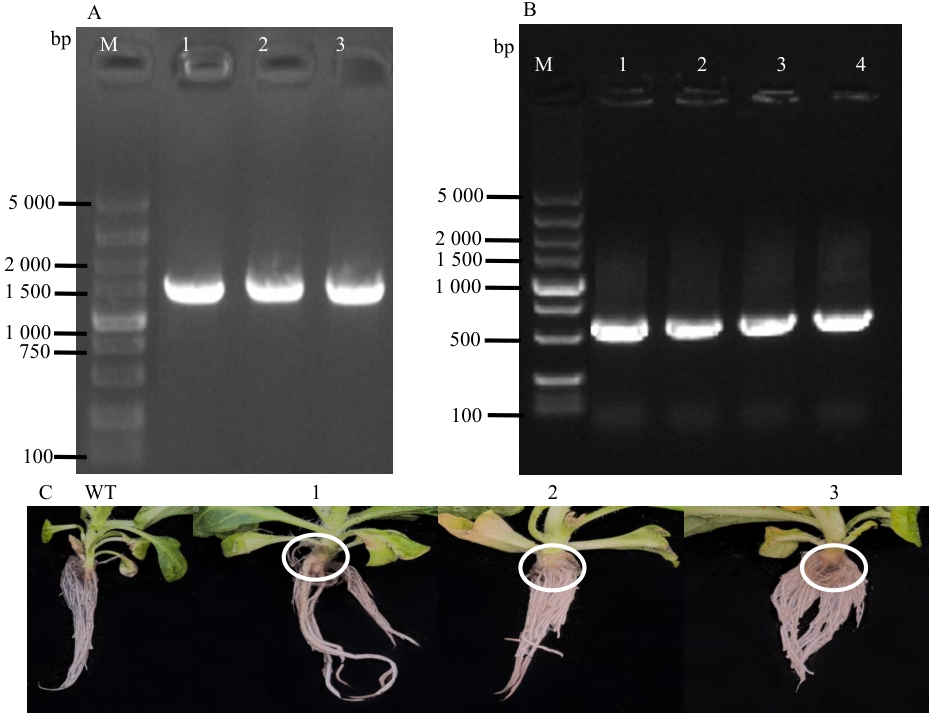
Fig. 8 Observation of GV3101 transformation, transgenic tobacco PCR, and transgenic plantA: 3302-bZIP29 transformed GV3101(M: DL5,000 DNA marker, 1-3: GV3101 colony). B: Transgenic tobacco PCR (M: DL5,000 DNA marker, 1-4: transgenic tobacco). C: Phenotype of transgenic tobacco (WT: wild type, 1-3: transgenic tobaccos)
| [1] | Weidemüller P, Kholmatov M, Petsalaki E, et al. Transcription factors: Bridge between cell signaling and gene regulation [J]. Proteomics, 2021, 21(23/24): 2000034. |
| [2] | Landschulz WH, Johnson PF, McKnight SL. The leucine zipper: a hypothetical structure common to a new class of DNA binding proteins [J]. Science, 1988, 240(4860): 1759-1764. |
| [3] | Zhang TT, Zhou Y. Plant transcription factors and salt stress [M]//Plant Transcription Factors. Academic Press, 2023: 369-381. |
| [4] | Kuravsky M, Grey G, Kelly C F, et al. A basic leucine zipper uses a dimer pathway to locate its targets in DNA mixtures[J]. bioRxiv, 2025: 2025.08. 25.672131. |
| [5] | Yu Y, Qian YC, Jiang MY, et al. Regulation mechanisms of plant basic leucine zippers to various abiotic stresses [J]. Front Plant Sci, 2020, 11: 1258. |
| [6] | Wang BX, Xu B, Liu Y, et al. A Novel mechanisms of the signaling cascade associated with the SAPK10-bZIP20-NHX1 synergistic interaction to enhance tolerance of plant to abiotic stress in rice (Oryza sativa L.) [J]. Plant Sci, 2022, 323: 111393. |
| [7] | Hussain Q, Asim M, Zhang R, et al. Transcription factors interact with ABA through gene expression and signaling pathways to mitigate drought and salinity stress [J]. Biomolecules, 2021, 11(8): 1159. |
| [8] | Yoshida T, Fujita Y, Sayama H, et al. AREB1, AREB2, and ABF3 are master transcription factors that cooperatively regulate ABRE-dependent ABA signaling involved in drought stress tolerance and require ABA for full activation [J]. Plant J, 2010, 61(4): 672-685. |
| [9] | Manna M, Thakur T, Chirom O, et al. Transcription factors as key molecular target to strengthen the drought stress tolerance in plants [J]. Physiol Plant, 2021, 172(2): 847-868. |
| [10] | 曹丽茹, 马晨晨, 庞芸芸, 等. 玉米bZIP转录因子ZmbZIP26的分子特征、亚细胞定位及响应非生物胁迫的表达分析 [J]. 华北农学报, 2023, 38(4): 1-10. |
| Cao LR, Ma CC, Pang YY, et al. Molecular characteristics, subcellular localization and expression analysis of maize bZIP transcription factor ZmbZIP26 in response to abiotic stress [J]. Acta Agric Boreali Sin, 2023, 38(4): 1-10. | |
| [11] | 郭纯, 宋桂梅, 闫艳, 等. 西洋参bZIP基因家族全基因组鉴定和表达分析 [J]. 生物技术通报, 2024, 40(4): 167-178. |
| Guo C, Song GM, Yan Y, et al. Genome wide identification and expression analysis of the bZIP gene family in Panax quinquefolius [J]. Biotechnol Bull, 2024, 40(4): 167-178. | |
| [12] | Gai WX, Ma X, Qiao YM, et al. Characterization of the bZIP transcription factor family in pepper (Capsicum annuum L.): CabZIP25 positively modulates the salt tolerance [J]. Front Plant Sci, 2020, 11: 139. |
| [13] | 张蕾, 陈昕涛, 白丽娟, 等. 大豆bZIP转录因子基因GmbZIP33的克隆与功能分析 [J]. 大豆科学, 2023, 42(5): 524-531. |
| Zhang L, Chen XT, Bai LJ, et al. Cloning and functional analysis of GmbZIP33 encoding bZIP transcription factor from soybean [J]. Soybean Sci, 2023, 42(5): 524-531. | |
| [14] | Li MY, Hwarari D, Li Y, et al. The bZIP transcription factors in Liriodendron chinense: Genome-wide recognition, characteristics and cold stress response [J]. Front Plant Sci, 2022, 13: 1035627. |
| [15] | Hsieh TH, Li CW, Su RC, et al. A tomato bZIP transcription factor, SlAREB, is involved in water deficit and salt stress response [J]. Planta, 2010, 231(6): 1459-1473. |
| [16] | 曾力旺, 张小燕, 林墁, 等. 木薯MebZIP27基因的克隆与原核表达分析 [J]. 福建农业学报, 2025, 40(1): 1-9. |
| Zeng LW, Zhang XY, Lin M, et al. Cloning and prokaryotic expression analysis of cassava MebZIP27 gene [J]. Fujian J Agric Sci, 2025, 40(1): 1-9. | |
| [17] | 曹晓宇, 王学敏, 郭继承, 等. 紫花苜蓿bZIP转录因子MsbZIP1的克隆、表达特性和遗传转化分析 [J]. 草地学报, 2020, 28(3): 613-622. |
| Cao XY, Wang XM, Guo JC, et al. Cloning, expression characteristics and genetic transformation analysis of a bZIP transcription factor MsbZIP1 in alfalfa [J]. Acta Agrestia Sin, 2020, 28(3): 613-622. | |
| [18] | Liu SX, Qin B, Fang QX, et al. Genome-wide identification, phylogeny and expression analysis of the bZIP gene family in Alfalfa (Medicago sativa) [J]. Biotechnol Biotechnol Equip, 2021, 35(1): 905-916. |
| [19] | Zhao K, Chen S, Yao WJ, et al. Genome-wide analysis and expression profile of the bZIP gene family in poplar [J]. BMC Plant Biol, 2021, 21(1): 122. |
| [20] | Dash M, Yordanov YS, Georgieva T, et al. Poplar PtabZIP1-like enhances lateral root formation and biomass growth under drought stress [J]. Plant J, 2017, 89(4): 692-705. |
| [21] | 赵安琪, 尹跃, 何军, 等. 枸杞LbaHY5基因克隆、亚细胞定位及表达分析 [J]. 华北农学报, 2024, 39(6): 76-83. |
| Zhao AQ, Yin Y, He J, et al. Cloning, subcellular localization and expression analysis of LbaHY5 gene from Lycium barbarum [J]. Acta Agric Boreali Sin, 2024, 39(6): 76-83. | |
| [22] | Yang SS, Zhang XX, Zhang XM, et al. A bZIP transcription factor, PqbZIP1, is involved in the plant defense response of American ginseng [J]. PeerJ, 2022, 10: e12939. |
| [23] | Niu XL, Fu DQ. The roles of BLH transcription factors in plant development and environmental response [J]. Int J Mol Sci, 2022, 23(7): 3731. |
| [24] | Verma S, Attuluri VPS, Robert HS. Transcriptional control of Arabidopsis seed development [J]. Planta, 2022, 255(4): 90. |
| [25] | Guo ZL, Dzinyela R, Yang LM, et al. bZIP transcription factors: structure, modification, abiotic stress responses and application in plant improvement [J]. Plants, 2024, 13(15): 2058. |
| [26] | Alonso-Blanco C, Aarts MGM, Bentsink L, et al. What has natural variation taught us about plant development, physiology, and adaptation? [J]. Plant Cell, 2009, 21(7): 1877-1896. |
| [27] | Li JS, Wang XM, Zhang YL, et al. cGMP regulates hydrogen peroxide accumulation in calcium-dependent salt resistance pathway in Arabidopsis thaliana roots [J]. Planta, 2011, 234(4): 709-722. |
| [28] | Kaur P, Lui C, Dudchenko O, et al. Delineating the Tnt1 insertion landscape of the model legume Medicago truncatula cv. R108 at the hi-C resolution using a chromosome-length genome assembly [J]. Int J Mol Sci, 2021, 22(9): 4326. |
| [29] | Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method [J]. Methods, 2001, 25(4): 402-408. |
| [30] | Riechmann JL, Heard J, Martin G, et al. Arabidopsis Transcription factors: genome-wide comparative analysis among eukaryotes [J]. Science, 2000, 290(5499): 2105-2110. |
| [31] | Fujii H, Verslues PE, Zhu JK. Identification of two protein kinases required for abscisic acid regulation of seed germination, root growth, and gene expression in Arabidopsis [J]. Plant Cell, 2007, 19(2): 485-494. |
| [32] | Yoshida T, Fujita Y, Maruyama K, et al. Four Arabidopsis AREB/ABF transcription factors function predominantly in gene expression downstream of SnRK2 kinases in abscisic acid signalling in response to osmotic stress [J]. Plant Cell Environ, 2015, 38(1): 35-49. |
| [33] | Fujii H, Zhu JK. Arabidopsis mutant deficient in 3 abscisic acid-activated protein kinases reveals critical roles in growth, reproduction, and stress [J]. Proc Natl Acad Sci U S A, 2009, 106(20): 8380-8385. |
| [34] | Cutler SR, Rodriguez PL, Finkelstein RR, et al. Abscisic acid: emergence of a core signaling network [J]. Annu Rev Plant Biol, 2010, 61: 651-679. |
| [35] | Luang S, Sornaraj P, Bazanova N, et al. The wheat TabZIP2 transcription factor is activated by the nutrient starvation-responsive SnRK3/CIPK protein kinase [J]. Plant Mol Biol, 2018, 96(6): 543-561. |
| [36] | Han XY, Wei XP, Lu WJ, et al. Transcriptional regulation of KCS gene by bZIP29 and MYB70 transcription factors during ABA-stimulated wound suberization of kiwifruit (Actinidia deliciosa) [J]. BMC Plant Biol, 2022, 22(1): 23. |
| [37] | Zhang LN, Zhang LC, Xia C, et al. A novel wheat bZIP transcription factor, TabZIP60, confers multiple abiotic stress tolerances in transgenic Arabidopsis [J]. Physiol Plant, 2015, 153(4): 538-554. |
| [38] | Van Leene J, Blomme J, Kulkarni SR, et al. Functional characterization of the Arabidopsis transcription factor bZIP29 reveals its role in leaf and root development [J]. J Exp Bot, 2016, 67(19): 5825-5840. |
| [1] | YANG Juan, FENG Hui, JI Nai-zhe, SUN Li-ping, WANG Yun, ZHANG Jia-nan, ZHAO Shi-wei. Cloning and Functional Analysis of AP2/ERF Transcription Factors RcERF4 and RcRAP2-12 in Rose [J]. Biotechnology Bulletin, 2026, 42(1): 150-160. |
| [2] | WU Cui-cui, CHEN Deng-ke, LAN Gang, XIA Zhi, LI Peng-bo. Bioinformatics Analysis of Peanut Transcription Factor AhHDZ70 and Its Tolerances to Salt and Drought [J]. Biotechnology Bulletin, 2026, 42(1): 198-207. |
| [3] | LIU Jia-li, SONG Jing-rong, ZHAO Wen-yu, ZHANG Xin-yuan, ZHAO Zi-yang, CAO Yi-bo, ZHANG Ling-yun. Identification of the R2R3-MYB Gene and Expression Analysis of Flavonoid Regulatory Genes in Blueberry [J]. Biotechnology Bulletin, 2025, 41(9): 124-138. |
| [4] | LI Yu-zhen, LI Meng-dan, ZHANG Wei, PENG Ting. Functional Study of RmEXPB2 Genein Rosa multiflora Based on the Identification of the Expansin Gene Family in Rosa sp. [J]. Biotechnology Bulletin, 2025, 41(9): 182-194. |
| [5] | ZHANG Chao-chao, HAN Kai-yuan, WANG Tong, CHEN Zhong. Cloning and Functional Analysis of PtoYABBY2 and PtoYABBY12 in Populus tomentosa [J]. Biotechnology Bulletin, 2025, 41(9): 256-264. |
| [6] | SHI Fa-chao, JIANG Yong-hua, LIU Hai-lun, WEN Ying-jie, YAN Qian. Cloning and Functional Analysis of LcTFL1 Gene in Litchi chinensis Sonn. [J]. Biotechnology Bulletin, 2025, 41(9): 159-167. |
| [7] | HUANG Shi-yu, TIAN Shan-shan, YANG Tian-wei, GAO Man-rong, ZHANG Shang-wen. Genome-wide Identification and Expression Pattern Analysis of WRI1 Gene Family in Erythropalum scandens [J]. Biotechnology Bulletin, 2025, 41(8): 242-254. |
| [8] | KANG Qin, WANG Xia, SHEN Ming-yang, XU Jing-tian, CHEN Shi-lan, LIAO Ping-yang, XU Wen-zhi, WU Wei, XU Dong-bei. Cloning and Expression Analysis of the UV-B Receptor Gene McUVR8 in Mentha canadensis L. [J]. Biotechnology Bulletin, 2025, 41(8): 255-266. |
| [9] | ZHAI Ying, JI Jun-jie, CHEN Jiong-xin, YU Hai-wei, LI Shan-shan, ZHAO Yan, MA Tian-yi. Heterologous Overexpression of Soybean GmNF-YB24 Improves the Resistance of Transgenic Tobacco to Drought [J]. Biotechnology Bulletin, 2025, 41(8): 137-145. |
| [10] | WANG Fang, QIAO Shuai, SONG Wei, CUI Peng-juan, LIAO An-zhong, TAN Wen-fang, YANG Song-tao. Genome-wide Identification of the IbNRT2 Gene Family and Its Expression in Sweet Potato [J]. Biotechnology Bulletin, 2025, 41(7): 193-204. |
| [11] | DONG Xu-kun, CHE Yong-mei, WANG Ming-shuo, LUO Zheng-gang, GUAN En-sen, ZHAO Fang-gui, YE Qing, LIU Xin. Effects of Co-treatment of Nano-silica and Bacillus cereus SS1 on the Growth of Tobacco [J]. Biotechnology Bulletin, 2025, 41(7): 292-298. |
| [12] | ZHANG Ze, YANG Xiu-li, NING Dong-xian. Identification of 4CL Gene Family in Arachis hypogaea L. and Expression Analysis in Response to Drought and Salt Stress [J]. Biotechnology Bulletin, 2025, 41(7): 117-127. |
| [13] | HUANG Xu-sheng, ZHOU Ya-li, CHAI Xu-dong, WEN Jing, WANG Ji-ping, JIA Xiao-yun, LI Run-zhi. Cloning of Plastidial PfLPAT1B Gene from Perilla frutescens and Its Functional Analysis in Oil Biosynthesis [J]. Biotechnology Bulletin, 2025, 41(7): 226-236. |
| [14] | XU Hui-zhen, SHANTWANA Ghimire, RAJU Kharel, YUE Yun, SI Huai-jun, TANG Xun. Analysis of the Potato SUMO E3 Ligase Gene Family and Cloning and Expression Pattern of StSIZ1 [J]. Biotechnology Bulletin, 2025, 41(6): 119-129. |
| [15] | YANG Chun, WANG Xiao-qian, WANG Hong-jun, CHAO Yue-hui. Cloning, Subcellular Localization and Expression Analysis of MtZHD4 Gene from Medicago truncatula [J]. Biotechnology Bulletin, 2025, 41(5): 244-254. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||