








Biotechnology Bulletin ›› 2025, Vol. 41 ›› Issue (12): 1-15.doi: 10.13560/j.cnki.biotech.bull.1985.2025-0637
FANG Yu-jie( ), LIU Kuan, CUI Han-bing, WANG You-ping
), LIU Kuan, CUI Han-bing, WANG You-ping
Received:2025-06-18
Online:2025-12-26
Published:2026-01-06
Contact:
FANG Yu-jie
E-mail:yjfang@yzu.edu.cn
FANG Yu-jie, LIU Kuan, CUI Han-bing, WANG You-ping. Recent Advances in Plant SUMO E3 Ligases[J]. Biotechnology Bulletin, 2025, 41(12): 1-15.
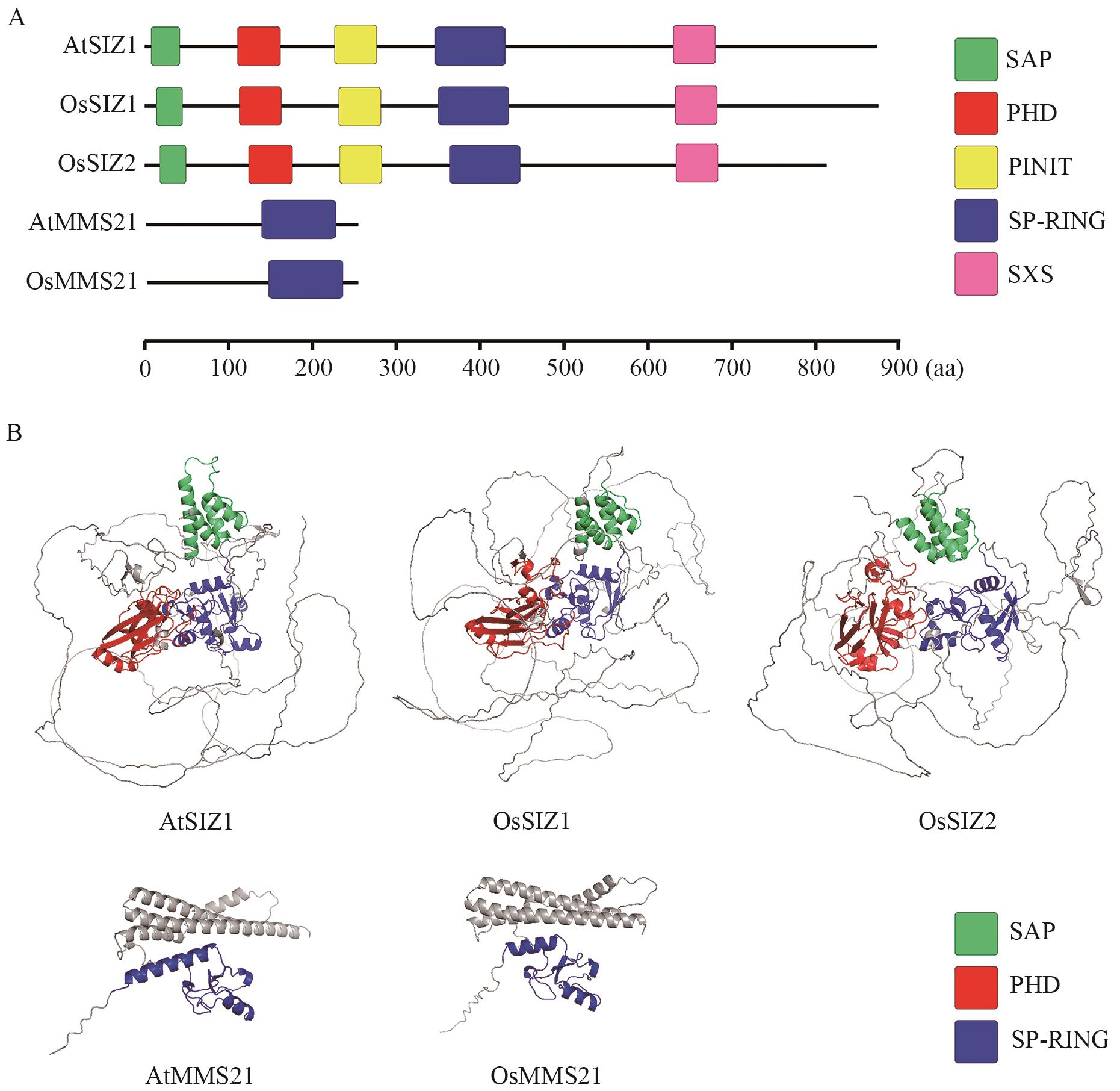
Fig. 1 Conserved domains of SUMO E3 ligases in Arabidopsis and riceA. Diagram of conserved domains of SUMO E3 ligases. B. Prediction of the tertiary structure of SUMO E3 ligases
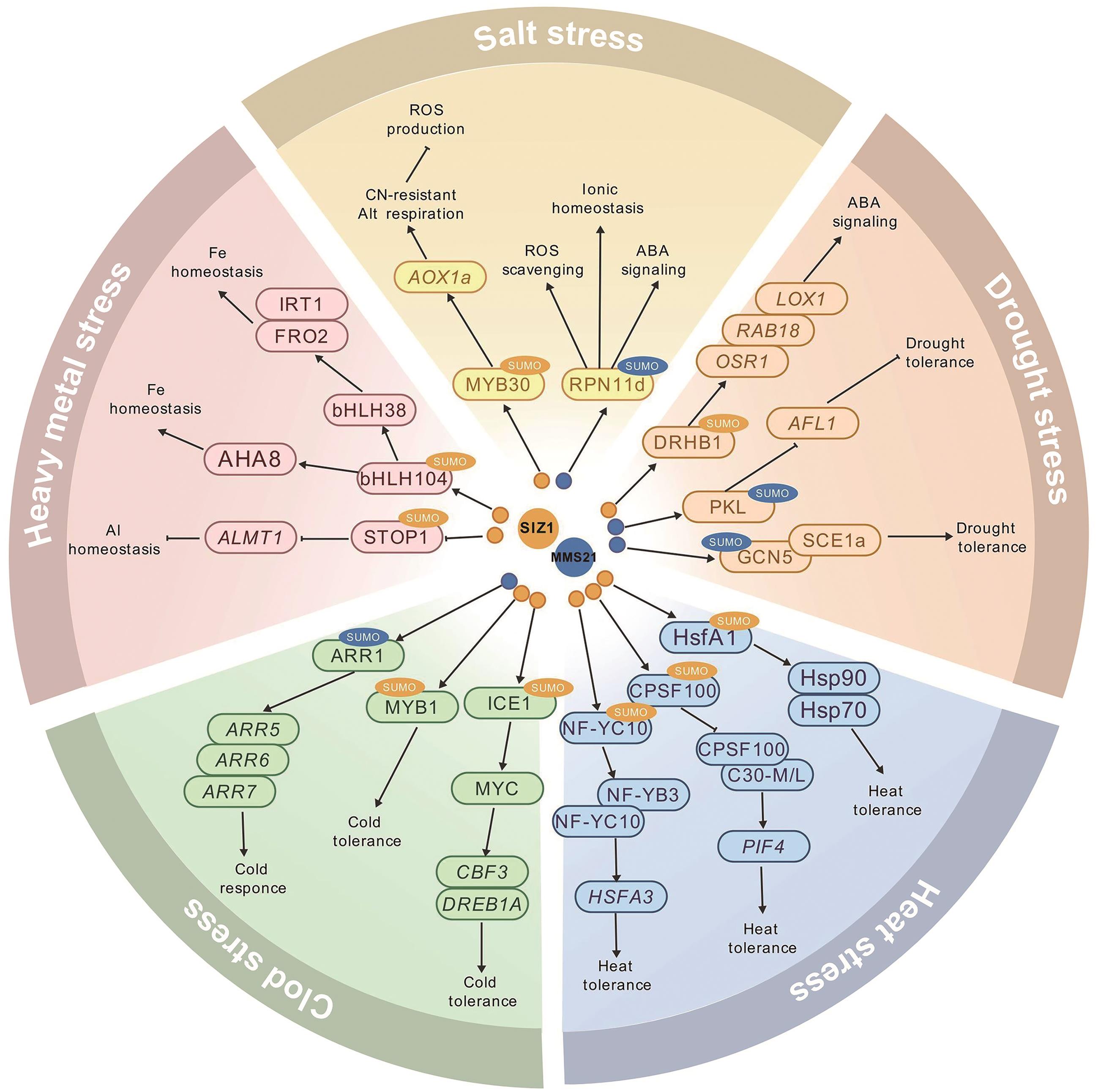
Fig. 2 The regulatory network of SUMO E3 ligase-mediated sumoylation in plant abiotic stress responses: SIZ1-mediated sumoylation; : MMS21-mediated sumoylation; →: positive regulation; : negative regulation
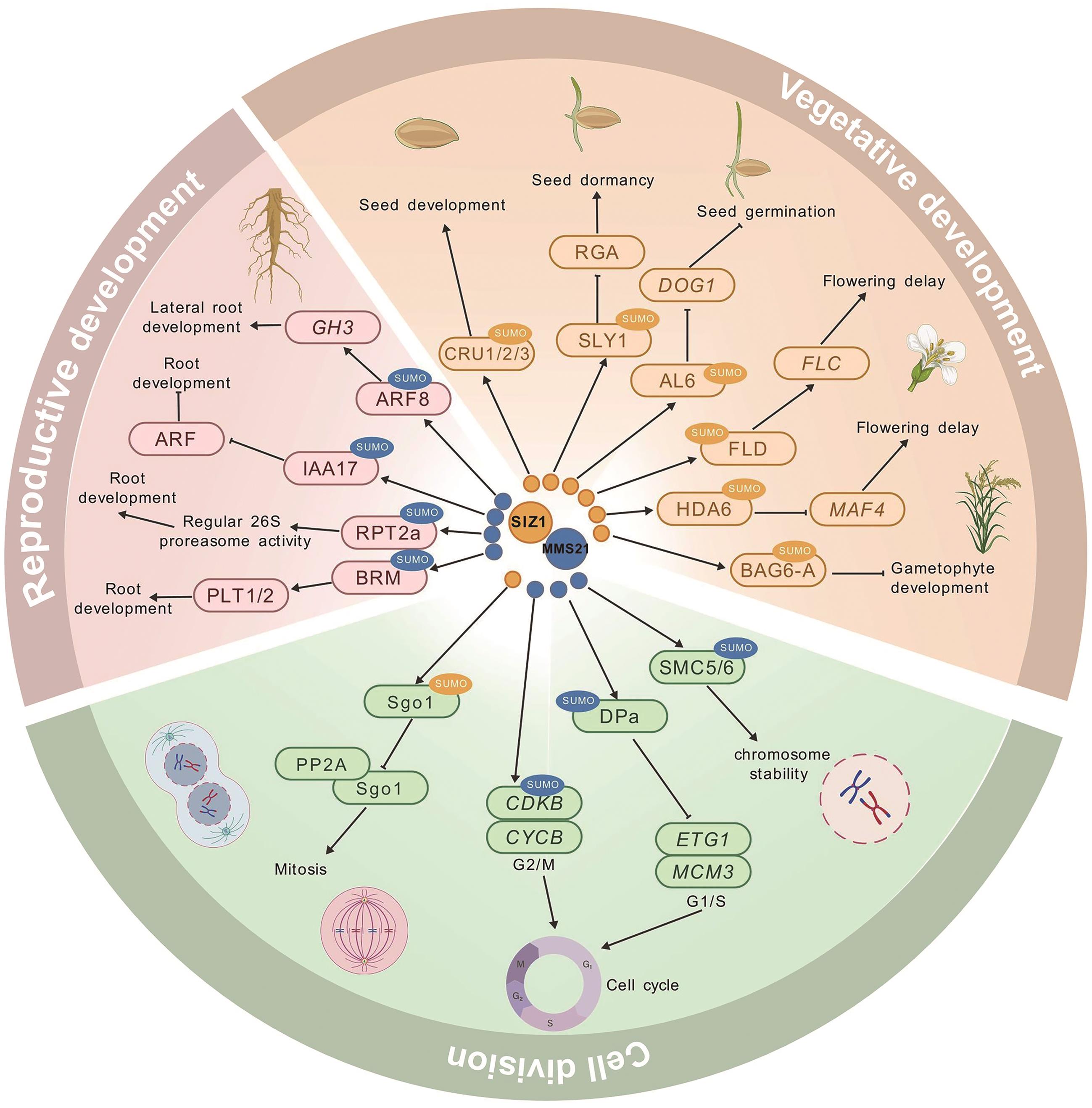
Fig. 3 Regulatory network of SUMO E3 ligase-mediated sumoylation in plant growth and development: SIZ1-mediated sumoylation; : MMS21-mediated sumoylation; →: positive regulation; : negative regulation
| [1] | 陈艳梅. 蛋白质翻译后修饰之间的互作关系及其协同调控机理 [J]. 生物技术通报, 2024, 40(2): 1-8. |
| Chen YM. Crosstalk between different post-translational modifications and its regulatory mechanisms in plant growth and development [J]. Biotechnol Bull, 2024, 40(2): 1-8. | |
| [2] | Li ZX, Qin LQ, Wang DJ, et al. Protein post-translational modification in plants: Regulation and beyond [J]. Hortic Plant J, 2025 |
| [3] | Benlloch R, Lois LM. Sumoylation in plants: mechanistic insights and its role in drought stress [J]. J Exp Bot, 2018, 69(19): 4539-4554. |
| [4] | Zhang ZX, Dong YJ, Wang XY, et al. Protein post-translational modifications (PTMs) unlocking resilience to abiotic stress in horticultural crops: a review [J]. Int J Biol Macromol, 2025, 306: 141772. |
| [5] | Elrouby N. Analysis of small ubiquitin-like modifier (SUMO) targets reflects the essential nature of protein SUMOylation and provides insight to elucidate the role of SUMO in plant development [J]. Plant Physiol, 2015, 169(2): 1006-1017. |
| [6] | Yau TY, Sander W, Eidson C, et al. SUMO interacting motifs: structure and function [J]. Cells, 2021, 10(11): 2825. |
| [7] | Jmii S, Cappadocia L. Plant SUMO E3 ligases: function, structural organization, and connection with DNA [J]. Front Plant Sci, 2021, 12: 652170. |
| [8] | Geoffroy MC, Hay RT. An additional role for SUMO in ubiquitin-mediated proteolysis [J]. Nat Rev Mol Cell Biol, 2009, 10(8): 564-568. |
| [9] | Vierstra RD. The expanding universe of ubiquitin and ubiquitin-like modifiers [J]. Plant Physiol, 2012, 160(1): 2-14. |
| [10] | Bernier-Villamor V, Sampson DA, Matunis MJ, et al. Structural basis for E2-mediated SUMO conjugation revealed by a complex between ubiquitin-conjugating enzyme Ubc9 and RanGAP1 [J]. Cell, 2002, 108(3): 345-356. |
| [11] | Johnson ES, Gupta AA. An E3-like factor that promotes SUMO conjugation to the yeast septins [J]. Cell, 2001, 106(6): 735-744. |
| [12] | Yunus AA, Lima CD. Structure of the siz/PIAS SUMO E3 ligase Siz1 and determinants required for SUMO modification of PCNA [J]. Mol Cell, 2009, 35(5): 669-682. |
| [13] | Pichler A, Gast A, Seeler JS, et al. The nucleoporin RanBP2 has SUMO1 E3 ligase activity [J]. Cell, 2002, 108(1): 109-120. |
| [14] | Kagey MH, Melhuish TA, Wotton D. The polycomb protein Pc2 is a SUMO E3 [J]. Cell, 2003, 113(1): 127-137. |
| [15] | Miura K, Rus A, Sharkhuu A, et al. The Arabidopsis SUMO E3 ligase SIZ1 controls phosphate deficiency responses [J]. Proc Natl Acad Sci U S A, 2005, 102(21): 7760-7765. |
| [16] | Huang LX, Yang SG, Zhang SC, et al. The Arabidopsis SUMO E3 ligase AtMMS21, a homologue of NSE2/MMS21, regulates cell proliferation in the root [J]. Plant J, 2009, 60(4): 666-678. |
| [17] | Cheong MS, Park HC, Bohnert HJ, et al. Structural and functional studies of SIZ1, a PIAS-type SUMO E3 ligase from Arabidopsis [J]. Plant Signal Behav, 2010, 5(5): 567-569. |
| [18] | Park HC, Kim H, Koo SC, et al. Functional characterization of the SIZ/PIAS-type SUMO E3 ligases, OsSIZ1 and OsSIZ2 in rice [J]. Plant Cell Environ, 2010, 33(11): 1923-1934. |
| [19] | Ishida T, Yoshimura M, Miura K, et al. MMS21/HPY2 and SIZ1, two Arabidopsis SUMO E3 ligases, have distinct functions in development [J]. PLoS One, 2012, 7(10): e46897. |
| [20] | Catala R, Ouyang J, Abreu IA, et al. The Arabidopsis E3 SUMO ligase SIZ1 regulates plant growth and drought responses [J]. Plant Cell, 2007, 19(9): 2952-2966. |
| [21] | Yoo CY, Miura K, Jin JB, et al. SIZ1 small ubiquitin-like modifier E3 ligase facilitates basal thermotolerance in Arabidopsis independent of salicylic acid [J]. Plant Physiol, 2006, 142(4): 1548-1558. |
| [22] | Zhou LJ, Li YY, Zhang RF, et al. The small ubiquitin-like modifier E3 ligase MdSIZ1 promotes anthocyanin accumulation by sumoylating MdMYB1 under low-temperature conditions in apple [J]. Plant Cell Environ, 2017, 40(10): 2068-2080. |
| [23] | Lee J, Nam J, Park HC, et al. Salicylic acid-mediated innate immunity in Arabidopsis is regulated by SIZ1 SUMO E3 ligase [J]. Plant J, 2007, 49(1): 79-90. |
| [24] | Miura K, Sato A, Ohta M, et al. Increased tolerance to salt stress in the phosphate-accumulating Arabidopsis mutants siz1 and pho2 [J]. Planta, 2011, 234(6): 1191-1199. |
| [25] | Mishra N, Srivastava AP, Esmaeili N, et al. Overexpression of the rice gene OsSIZ1 in Arabidopsis improves drought-, heat-, and salt-tolerance simultaneously [J]. PLoS One, 2018, 13(8): e0201716. |
| [26] | Han GL, Yuan F, Guo JR, et al. AtSIZ1 improves salt tolerance by maintaining ionic homeostasis and osmotic balance in Arabidopsis [J]. Plant Sci, 2019, 285: 55-67. |
| [27] | Gong QY, Li S, Zheng Y, et al. SUMOylation of MYB30 enhances salt tolerance by elevating alternative respiration via transcriptionally upregulating AOX1a in Arabidopsis [J]. Plant J, 2020, 102(6): 1157-1171. |
| [28] | Lei SK, Wang QZ, Chen Y, et al. Capsicum SIZ1 contributes to ABA-induced SUMOylation in pepper [J]. Plant Sci, 2022, 314: 111099. |
| [29] | Wang SB, Guo JS, Peng YY, et al. GmRPN11d positively regulates plant salinity tolerance by improving protein stability through SUMOylation [J]. Int J Biol Macromol, 2025, 294: 139393. |
| [30] | Joo H, Lim CW, Lee SC. Pepper SUMO E3 ligase CaDSIZ1 enhances drought tolerance by stabilizing the transcription factor CaDRHB1 [J]. New Phytol, 2022, 235(6): 2313-2330. |
| [31] | Zhang SC, Qi YL, Liu M, et al. SUMO E3 ligase AtMMS21 regulates drought tolerance in Arabidopsis thaliana [J]. J Integr Plant Biol, 2013, 55(1): 83-95. |
| [32] | Castro PH, Couto D, Freitas S, et al. SUMO proteases ULP1c and ULP1d are required for development and osmotic stress responses in Arabidopsis thaliana [J]. Plant Mol Biol, 2016, 92(1-2): 143-159. |
| [33] | Jing YX, Yang ZY, Yang RZ, et al. PKL is stabilized by MMS21 to negatively regulate Arabidopsis drought tolerance through directly repressing AFL1 transcription [J]. New Phytol, 2023, 239(3): 920-935. |
| [34] | Feng TY, Wang YX, Zhang MC, et al. ZmSCE1a positively regulates drought tolerance by enhancing the stability of ZmGCN5 [J]. Plant J, 2024, 120(5): 2101-2112. |
| [35] | Hammoudi V, Fokkens L, Beerens B, et al. The Arabidopsis SUMO E3 ligase SIZ1 mediates the temperature dependent trade-off between plant immunity and growth [J]. PLoS Genet, 2018, 14(1): e1007157. |
| [36] | Rytz TC, Miller MJ, McLoughlin F, et al. SUMOylome profiling reveals a diverse array of nuclear targets modified by the SUMO ligase SIZ1 during heat stress [J]. Plant Cell, 2018, 30(5): 1077-1099. |
| [37] | Zhang S, Wang SJ, Lv JL, et al. SUMO E3 ligase SlSIZ1 facilitates heat tolerance in tomato [J]. Plant Cell Physiol, 2018, 59(1): 58-71. |
| [38] | Yu ZB, Wang J, Zhang C, et al. SIZ1-mediated SUMOylation of CPSF100 promotes plant thermomorphogenesis by controlling alternative polyadenylation [J]. Mol Plant, 2024, 17(9): 1392-1406. |
| [39] | Zheng XT, Wang CJ, Lin WX, et al. Importation of chloroplast proteins under heat stress is facilitated by their SUMO conjugations [J]. New Phytol, 2022, 235(1): 173-187. |
| [40] | Huang JW, Huang JJ, Feng QY, et al. SUMOylation facilitates the assembly of a Nuclear Factor-Y complex to enhance thermotolerance in Arabidopsis [J]. J Integr Plant Biol, 2023, 65(3): 692-702. |
| [41] | Kwak JS, Song JT, Seo HS. E3 SUMO ligase SIZ1 splicing variants localize and function according to external conditions [J]. Plant Physiol, 2024, 195(2): 1601-1623. |
| [42] | Miura K, Jin JB, Lee J, et al. SIZ1-mediated sumoylation of ICE1 controls CBF3/DREB1A expression and freezing tolerance in Arabidopsis [J]. Plant Cell, 2007, 19(4): 1403-1414. |
| [43] | Miura K, Ohta M. SIZ1, a small ubiquitin-related modifier ligase, controls cold signaling through regulation of salicylic acid accumulation [J]. J Plant Physiol, 2010, 167(7): 555-560. |
| [44] | Kim JY, Jang IC, Seo HS. COP1 controls abiotic stress responses by modulating AtSIZ1 function through its E3 ubiquitin ligase activity [J]. Front Plant Sci, 2016, 7: 1182. |
| [45] | Jiang H, Zhou LJ, Gao HN, et al. The transcription factor MdMYB2 influences cold tolerance and anthocyanin accumulation by activating SUMO E3 ligase MdSIZ1 in apple [J]. Plant Physiol, 2022, 189(4): 2044-2060. |
| [46] | Kang NY, Kim MJ, Jeong S, et al. HIGH PLOIDY2-mediated SUMOylation of transcription factor ARR1 controls two-component signaling in Arabidopsis [J]. Plant Cell, 2024, 36(9): 3521-3542. |
| [47] | Datta M, Kaushik S, Jyoti A, et al. SIZ1-mediated SUMOylation during phosphate homeostasis in plants: Looking beyond the tip of the iceberg [J]. Semin Cell Dev Biol, 2018, 74: 123-132. |
| [48] | Kenji Miura JL. SIZ1 regulation of phosphate starvation-induced root architecture remodeling involves the control of auxin accumulation [J]. Plant Physiol, 2011, 155(2): 1000-1012. |
| [49] | Pei WX, Jain A, Sun YF, et al. OsSIZ2 exerts regulatory influences on the developmental responses and phosphate homeostasis in rice [J]. Sci Rep, 2017, 7: 12280. |
| [50] | Wang HD, Sun R, Cao Y, et al. OsSIZ1, a SUMO E3 ligase gene, is involved in the regulation of the responses to phosphate and nitrogen in rice [J]. Plant Cell Physiol, 2015, 56(12): 2381-2395. |
| [51] | Zhang RF, Zhou LJ, Li YY, et al. Apple SUMO E3 ligase MdSIZ1 is involved in the response to phosphate deficiency [J]. J Plant Physiol, 2019, 232: 216-225. |
| [52] | Fang Q, Zhang J, Yang DL, et al. The SUMO E3 ligase SIZ1 partially regulates STOP1 SUMOylation and stability in Arabidopsis thaliana [J]. Plant Signal Behav, 2021, 16(5): 1899487. |
| [53] | Xu JM, Zhu JY, Liu JJ, et al. SIZ1 negatively regulates aluminum resistance by mediating the STOP1-ALMT1 pathway in Arabidopsis [J]. J Integr Plant Biol, 2021, 63(6): 1147-1160. |
| [54] | Park BS, Song JT, Seo HS. Arabidopsis nitrate reductase activity is stimulated by the E3 SUMO ligase AtSIZ1 [J]. Nat Commun, 2011, 2: 400. |
| [55] | Kim JY, Park BS, Park SW, et al. Nitrate reductases are relocalized to the nucleus by AtSIZ1 and their levels are negatively regulated by COP1 and ammonium [J]. Int J Mol Sci, 2018, 19(4): 1202. |
| [56] | Park BS, Kim SI, Song JT, et al. Arabidopsis SIZ1 positively regulates alternative respiratory bypass pathways [J]. BMB Rep, 2012, 45(6): 342-347. |
| [57] | Youn K, Jung H, Kim SI, et al. Arabidopsis CMT3 activity is positively regulated by AtSIZ1-mediated sumoylation [J]. Plant Sci, 2015, 239: 209-215. |
| [58] | Castro PH, Verde N, Lourenço T, et al. SIZ1-dependent post-translational modification by SUMO modulates sugar signaling and metabolism in Arabidopsis thaliana [J]. Plant Cell Physiol, 2015, 56(12): 2297-2311. |
| [59] | Castro PH, Verde N, Tavares RM, et al. Sugar signaling regulation by Arabidopsis SIZ1-driven sumoylation is independent of salicylic acid [J]. Plant Signal Behav, 2018, 13(4): e1179417. |
| [60] | Castro PH, Verde N, Azevedo H. Arabidopsis thaliana growth is independently controlled by the SUMO E3 ligase SIZ1 and Hexokinase 1 [J]. MicroPubl Biol, 2020, (2020). |
| [61] | Rawat SS, Sandhya S, Laxmi A. Complex genetic interaction between glucose sensor HXK1 and E3 SUMO ligase SIZ1 in regulating plant morphogenesis [J]. Plant Signal Behav, 2024, 19(1): 2341506. |
| [62] | Chen CC, Chen YY, Tang IC, et al. Arabidopsis SUMO E3 ligase SIZ1 is involved in excess copper tolerance [J]. Plant Physiol, 2011, 156(4): 2225-2234. |
| [63] | Chen CC, Chen YY, Yeh KC. Effect of Cu content on the activity of Cu/ZnSOD1 in the Arabidopsis SUMO E3 ligase siz1 mutant [J]. Plant Signal Behav, 2011, 6(10): 1428-1430. |
| [64] | Chen CC, Chien WF, Lin NC, et al. Alternative functions of Arabidopsis Yellow STRIPE-LIKE3: from metal translocation to pathogen defense [J]. PLoS One, 2014, 9(5): e98008. |
| [65] | Zhan EB, Zhou HP, Li S, et al. OTS1-dependent deSUMOylation increases tolerance to high copper levels in Arabidopsis [J]. J Integr Plant Biol, 2018, 60(4): 310-322. |
| [66] | Zheng Z, Liu D. SIZ1 regulates phosphate deficiency-induced inhibition of primary root growth of Arabidopsis by modulating Fe accumulation and ROS production in its roots [J]. Plant Signal Behav, 2021, 16(10): 1946921. |
| [67] | Zhou LJ, Zhang CL, Zhang RF, et al. The SUMO E3 ligase MdSIZ1 targets MdbHLH104 to regulate plasma membrane H+-ATPase activity and iron homeostasis [J]. Plant Physiol, 2019, 179(1): 88-106. |
| [68] | Kenji Miura JL. Sumoylation of ABI5 by the Arabidopsis SUMO E3 ligase SIZ1 negatively regulates abscisic acid signaling [J]. Proc Natl Acad Sci U S A, 2009, 106(13): 5418-5423. |
| [69] | Zhang JJ, Lai JB, Wang FG, et al. A SUMO ligase AtMMS21 regulates the stability of the chromatin remodeler BRAHMA in root development [J]. Plant Physiol, 2017, 173(3): 1574-1582. |
| [70] | Yu MY, Meng BL, Wang FG, et al. A SUMO ligase AtMMS21 regulates activity of the 26S proteasome in root development [J]. Plant Sci, 2019, 280: 314-320. |
| [71] | Zhang CL, Wang GL, Zhang YL, et al. Apple SUMO E3 ligase MdSIZ1 facilitates SUMOylation of MdARF8 to regulate lateral root formation [J]. New Phytol, 2021, 229(4): 2206-2222. |
| [72] | Jiang JM, Xie Y, Du JJ, et al. A SUMO ligase OsMMS21 regulates rice development and auxin response [J]. J Plant Physiol, 2021, 263: 153447. |
| [73] | Zhang C, Yang Y, Yu ZB, et al. SUMO E3 ligase AtMMS21-dependent SUMOylation of AUXIN/INDOLE-3-ACETIC ACID 17 regulates auxin signaling [J]. Plant Physiol, 2023, 191(3): 1871-1883. |
| [74] | Zhang LE, Han Q, Xiong JW, et al. Sumoylation of BRI1-EMS-SUPPRESSOR 1 (BES1) by the SUMO E3 ligase SIZ1 negatively regulates brassinosteroids signaling in Arabidopsis thaliana [J]. Plant Cell Physiol, 2019, 60(10): 2282-2292. |
| [75] | Xiong JW, Yang FB, Wei F, et al. Inhibition of SIZ1-mediated SUMOylation of HOOKLESS1 promotes light-induced apical hook opening in Arabidopsis [J]. Plant Cell, 2023, 35(6): 2027-2043. |
| [76] | Zhao FL, Liu LF, Du JK, et al. BAG6-A from Fragaria viridis pollen modulates gametophyte development in diploid strawberry [J]. Plant Sci, 2023, 330: 111667. |
| [77] | Soo K, Kim SI, Woo P, et al. E3 SUMO ligase AtSIZ1 regulates the cruciferin content of Arabidopsis seeds [J]. Biochem Biophys Res Commun, 2019, 519(4): 761-766. |
| [78] | Kim SI, Park BS, Kim DY, et al. E3 SUMO ligase AtSIZ1 positively regulates SLY1-mediated GA signalling and plant development [J]. Biochem J, 2015, 469(2): 299-314. |
| [79] | Kim SI, Kwak JS, Song JT, et al. The E3 SUMO ligase AtSIZ1 functions in seed germination in Arabidopsis [J]. Physiol Plant, 2016, 158(3): 256-271. |
| [80] | Jing H, Liu W, Qu GP, et al. SUMOylation of AL6 regulates seed dormancy and thermoinhibition in Arabidopsis [J]. New Phytol, 2025, 245(3): 1040-1055. |
| [81] | Thangasamy S, Guo CL, Chuang MH, et al. Rice SIZ1, a SUMO E3 ligase, controls spikelet fertility through regulation of anther dehiscence [J]. New Phytol, 2011, 189(3): 869-882. |
| [82] | Liu C, Yu HS, Li LG. SUMO modification of LBD30 by SIZ1 regulates secondary cell wall formation in Arabidopsis thaliana [J]. PLoS Genet, 2019, 15(1): e1007928. |
| [83] | Son GH, Park BS, Song JT, et al. FLC-mediated flowering repression is positively regulated by sumoylation [J]. J Exp Bot, 2014, 65(1): 339-351. |
| [84] | Jin JB, Jin YH, Lee J, et al. The SUMO E3 ligase, AtSIZ1, regulates flowering by controlling a salicylic acid-mediated floral promotion pathway and through affects on FLC chromatin structure [J]. Plant J, 2008, 53(3): 530-540. |
| [85] | Kwak JS, Son GH, Kim SI, et al. Arabidopsis HIGH PLOIDY2 sumoylates and stabilizes flowering locus C through its E3 ligase activity [J]. Front Plant Sci, 2016, 7: 530. |
| [86] | Gao SJ, Zeng XQ, Wang JH, et al. Arabidopsis SUMO E3 ligase SIZ1 interacts with HDA6 and negatively regulates HDA6 function during flowering [J]. Cells, 2021, 10(11): 3001. |
| [87] | Xu PL, Yuan DK, Liu M, et al. AtMMS21, an SMC5/6 complex subunit, is involved in stem cell niche maintenance and DNA damage responses in Arabidopsis roots [J]. Plant Physiol, 2013, 161(4): 1755-1768. |
| [88] | Liu M, Shi SF, Zhang SC, et al. SUMO E3 ligase AtMMS21 is required for normal meiosis and gametophyte development in Arabidopsis [J]. BMC Plant Biol, 2014, 14(1): 153. |
| [89] | Zhang JY, Augustine RC, Suzuki M, et al. The SUMO ligase MMS21 profoundly influences maize development through its impact on genome activity and stability [J]. PLoS Genet, 2021, 17(10): e1009830. |
| [90] | Yoshimura M, Ishida T. Generation of viable hypomorphic and null mutant plants via CRISPR-Cas9 targeting mRNA splicing sites [J]. J Plant Res, 2025, 138(1): 189-196. |
| [91] | Liu YY, Lai JB, Yu MY, et al. The Arabidopsis SUMO E3 ligase AtMMS21 dissociates the E2Fa/DPa complex in cell cycle regulation [J]. Plant Cell, 2016, 28(9): 2225-2237. |
| [92] | Su XB, Wang ML, Schaffner C, et al. SUMOylation stabilizes sister kinetochore biorientation to allow timely anaphase [J]. J Cell Biol, 2021, 220(7): e202005130. |
| [93] | Wang GL, Zhang CL, Huo HQ, et al. The SUMO E3 ligase MdSIZ1 sumoylates a cell number regulator MdCNR8 to control organ size [J]. Front Plant Sci, 2022, 13: 836935. |
| [94] | Yuan MH, Ngou BPM, Ding PT, et al. PTI-ETI crosstalk: an integrative view of plant immunity [J]. Curr Opin Plant Biol, 2021, 62: 102030. |
| [95] | Zhang Y, Zeng LR. Crosstalk between ubiquitination and other post-translational protein modifications in plant immunity [J]. Plant Commun, 2020, 1(4): 100041. |
| [96] | Fu ZQ, Dong XN. Systemic acquired resistance: turning local infection into global defense [J]. Annu Rev Plant Biol, 2013, 64: 839-863. |
| [97] | Gou MY, Huang QS, Qian WQ, et al. Sumoylation E3 ligase SIZ1 modulates plant immunity partly through the immune receptor gene SNC1 in Arabidopsis [J]. Mol Plant Microbe Interact, 2017, 30(4): 334-342. |
| [98] | Niu D, Lin XL, Kong XX, et al. SIZ1-mediated SUMOylation of TPR1 suppresses plant immunity in Arabidopsis [J]. Mol Plant, 2019, 12(2): 215-228. |
| [99] | Xie B, Luo MY, Li QY, et al. NUA positively regulates plant immunity by coordination with ESD4 to deSUMOylate TPR1 in Arabidopsis [J]. New Phytol, 2024, 241(1): 363-377. |
| [100] | Liu S, Lenoir CJG, Amaro TMMM, et al. Virulence strategies of an insect herbivore and oomycete plant pathogen converge on host E3 SUMO ligase SIZ1 [J]. New Phytol, 2022, 235(4): 1599-1614. |
| [101] | Hou SG, Liu ZY, Shen HX, et al. Damage-associated molecular pattern-triggered immunity in plants [J]. Front Plant Sci, 2019, 10: 646. |
| [102] | Zhang C, Wu YY, Liu JE, et al. SUMOylation controls peptide processing to generate damage-associated molecular patterns in Arabidopsis [J]. Dev Cell, 2025, 60(5): 696-705.e4. |
| [103] | Orosa B, Yates G, Verma V, et al. SUMO conjugation to the pattern recognition receptor FLS2 triggers intracellular signalling in plant innate immunity [J]. Nat Commun, 2018, 9: 5185. |
| [104] | Shi XX, Du YX, Li SJ, et al. The role of SUMO E3 ligases in signaling pathway of cancer cells [J]. Int J Mol Sci, 2022, 23(7): 3639. |
| [105] | Sang T, Xu YP, Qin GC, et al. Highly sensitive site-specific SUMOylation proteomics in Arabidopsis [J]. Nat Plants, 2024, 10(9): 1330-1342. |
| [106] | Augustine RC, York SL, Rytz TC, et al. Defining the SUMO system in maize: SUMOylation is up-regulated during endosperm development and rapidly induced by stress [J]. Plant Physiol, 2016, 171(3): 2191-2210. |
| [107] | Clark L, Sue-Ob K, Mukkawar V, et al. Understanding SUMO-mediated adaptive responses in plants to improve crop productivity [J]. Essays Biochem, 2022, 66(2): 155-168. |
| [1] | WANG Cong-huan, WU Guo-qiang, WEI Ming. Functional Mechanism of Plant CBL in Regulating the Responses to Abiotic and Biotic Stresses [J]. Biotechnology Bulletin, 2025, 41(7): 1-16. |
| [2] | WEI Yu-jia, LI Yan, KANG Yu-han, GONG Xiao-nan, DU Min, TU Lan, SHI Peng, YU Zi-han, SUN Yan, ZHANG Kun. Cloning and Expression Analysis of the CrMYB4 Gene in Carex rigescens [J]. Biotechnology Bulletin, 2025, 41(7): 248-260. |
| [3] | ZAN Shu-wen, XIE Huan-huan, ZHANG Yu-qin, WANG Wen-Juan, ZHANG Peng-fei, LIANG Jin-jun, WEN Peng-fei. VvAGAMOUS Regulates Carpel Development through VvCRABS CLAW in Grape [J]. Biotechnology Bulletin, 2025, 41(5): 208-217. |
| [4] | WANG Bin, LIN Wei, XIAO Yan-hui, YUAN Xiao. Research Progress in the Roles of Plant Glycine-rich Protein Family [J]. Biotechnology Bulletin, 2025, 41(2): 1-17. |
| [5] | WU Ding-jie, CHEN Ying-ying, XU Jing, LIU Yuan, ZHANG Hang, LI Rui-li. Research Progress in Plant Gibberellin Oxidase and Its Functions [J]. Biotechnology Bulletin, 2024, 40(7): 43-54. |
| [6] | CHEN Ying-ying, WU Ding-jie, LIU Yuan, ZHANG Hang, LIU Yan-jiao, WANG Jing-yu, LI Rui-li. Recent Advances in 14-3-3 Proteins and Their Functions in Plant [J]. Biotechnology Bulletin, 2024, 40(4): 12-22. |
| [7] | FU Wei, WEI Su-yun, CHEN Ying-nan. Research Progress in the Dynamic QTL Analysis of Plant Growth and Development [J]. Biotechnology Bulletin, 2024, 40(2): 9-19. |
| [8] | HOU Ying-xiang, FEI Si-tian, SONG Song-quan, LUO Yong, ZHANG Chao. Research Progress in MADS-box Family in Rice [J]. Biotechnology Bulletin, 2024, 40(11): 103-112. |
| [9] | GENG Ruo-han, WANG Bing-he, XU Chang-wen, QIAN Hong-ping, LIN Jin-xing, CUI Ya-ning. Research Progress in the Regulation of Protein Post-translational Modification in Plant Vesicle Transport [J]. Biotechnology Bulletin, 2024, 40(10): 139-148. |
| [10] | CHEN Meng-jiao, LI Yang-yang, WU Qian. Research Advances in Plant Growth and Stress Response Regulation Mediated by Glutamate Receptor-like Proteins [J]. Biotechnology Bulletin, 2024, 40(10): 62-75. |
| [11] | BI Fang-ling, ZHAO Shuang, LI Bin, LI Ai-qin, ZHANG Jian-heng, HE Pei-min. Research Progresses and Application in the Growth-promoting Effect of Symbiotic and Epiphytic Bacteria on Green Tide-causing Ulva prolifera [J]. Biotechnology Bulletin, 2024, 40(1): 32-44. |
| [12] | HU Hai-lin, XU Li, LI Xiao-xu, WANG Chen-can, MEI Man, DING Wen-jing, ZHAO Yuan-yuan. Advances in the Regulation of Plant Growth, Development and Stress Physiology by Small Peptide Hormones [J]. Biotechnology Bulletin, 2023, 39(7): 13-25. |
| [13] | FENG Shan-shan, WANG Lu, ZHOU Yi, WANG You-ping, FANG Yu-jie. Research Progresses on WOX Family Genes in Regulating Plant Development and Abiotic Stress Response [J]. Biotechnology Bulletin, 2023, 39(5): 1-13. |
| [14] | XUE Jiao ZHU Qing-feng FENG Yan-zhao CHEN Pei LIU Wen-hua ZHANG Ai-xia LIU Qin-jian ZHANG Qi YU Yang. Advances in Upstream Open Reading Frame in Plant Genes [J]. Biotechnology Bulletin, 2023, 39(4): 157-165. |
| [15] | WEI Ming WANG Xin-yu WU Guo-qiang ZHAO Meng. The Role of NAD-dependent Deacetylase SRT in Plant Epigenetic Inheritance Regulation [J]. Biotechnology Bulletin, 2023, 39(4): 59-70. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||