








Biotechnology Bulletin ›› 2026, Vol. 42 ›› Issue (4): 83-91.doi: 10.13560/j.cnki.biotech.bull.1985.2025-1073
Previous Articles Next Articles
LI Ya-qi1,2( ), SUN Meng1, LI Xiu-li1, WEI Jing-na1, ZHAO Lin-lin1, ZHAO Yun-ping1, LIU Zheng-hui1(
), SUN Meng1, LI Xiu-li1, WEI Jing-na1, ZHAO Lin-lin1, ZHAO Yun-ping1, LIU Zheng-hui1( ), SU Fan1(
), SU Fan1( )
)
Received:2025-10-10
Online:2026-02-09
Published:2026-02-09
Contact:
LIU Zheng-hui, SU Fan
E-mail:1072636135@qq.com;liuzhenghui_2025@126.com;fansu.agroscience@hotmail.com
LI Ya-qi, SUN Meng, LI Xiu-li, WEI Jing-na, ZHAO Lin-lin, ZHAO Yun-ping, LIU Zheng-hui, SU Fan. Optimization of a High-performance and Low-cost Fluorescence Detection Buffer with Broad Compatibility across Cas12a Orthologs[J]. Biotechnology Bulletin, 2026, 42(4): 83-91.
| 名称 Name | 序列 Sequences(5'-3') |
|---|---|
| dsDNA activator | CTCGCGAGTCCCAACACCAAGCTGGGCTTGAGGGTTGAAATGACGCTCGAACAGGCATGCCCGCCAGAATACTGGCGGGCGCAATGTGCGTTCAAAGATTCGATGATTCACTGAATTCTGCAATTCACATTACTTATCGCATTTT GCTGCGTTCTTCATCGATGC CAGAACCAAGAGATCCGTTGTTGAAAGTTTTGATTTATTTATGGTTTTACTCAGAAGTTACATATAGAAACAGAGTTTAGGGGTCCTCTGGCGGGCCGTCCCGTTTTACCGGGAGCGGGCTGATCCGCCGAGGCA |
| crRNA-F | TAATACGACTCACTATAGGGAATTTCTACTGTTGTAGAT |
| crRNA-R | GCATCGATGAAGAACGCAGCATCTACAACAGTAGAAATT |
| crRNA |
Table 1 dsDNA activator fragments, crRNA sequences, and primer information used in the Cas12a system
| 名称 Name | 序列 Sequences(5'-3') |
|---|---|
| dsDNA activator | CTCGCGAGTCCCAACACCAAGCTGGGCTTGAGGGTTGAAATGACGCTCGAACAGGCATGCCCGCCAGAATACTGGCGGGCGCAATGTGCGTTCAAAGATTCGATGATTCACTGAATTCTGCAATTCACATTACTTATCGCATTTT GCTGCGTTCTTCATCGATGC CAGAACCAAGAGATCCGTTGTTGAAAGTTTTGATTTATTTATGGTTTTACTCAGAAGTTACATATAGAAACAGAGTTTAGGGGTCCTCTGGCGGGCCGTCCCGTTTTACCGGGAGCGGGCTGATCCGCCGAGGCA |
| crRNA-F | TAATACGACTCACTATAGGGAATTTCTACTGTTGTAGAT |
| crRNA-R | GCATCGATGAAGAACGCAGCATCTACAACAGTAGAAATT |
| crRNA |
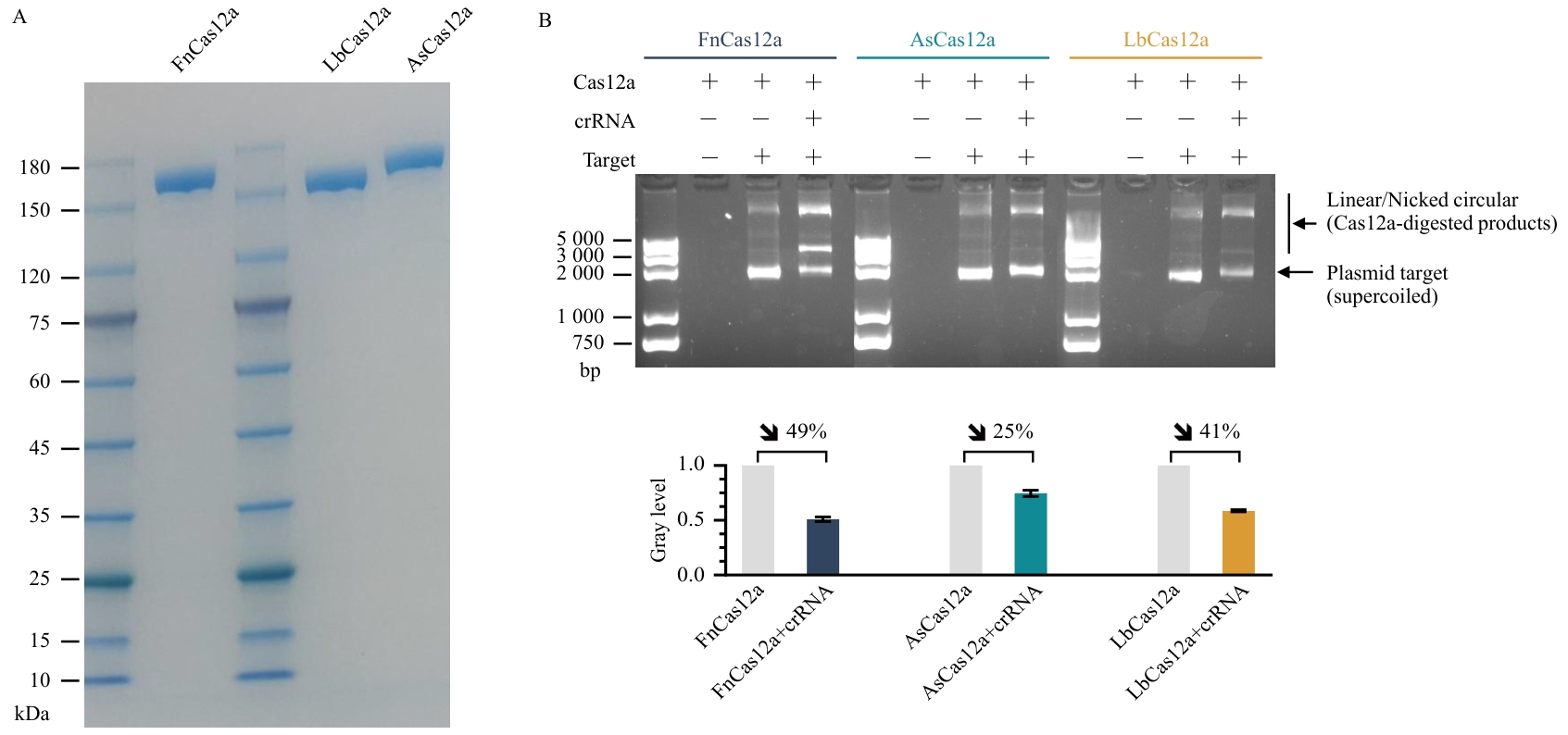
Fig. 1 Expression, purification, and in vitro cleavage activity analysis of Cas12a proteinsA: SDS-PAGE analysis of recombinant Cas12a proteins; B: In vitro DNA cleavage assays of three Cas12a proteins; Top: Agarose gel electrophoresis showing crRNA-guided cleavage of plasmid DNA substrates by each Cas12a protein. Bottom: Quantitative grayscale analysis of plasmid targets (n=3). Data are presented as mean ± SEM.

Fig. 2 Effect of key reaction conditions for the CRISPR/Cas12a fluorescence biosensor performanceA: Effect of pH; B: Effect of Tris-HCl concentration; C: Effect of Ca²⁺ concentration; D: Effect of Mg²⁺ concentration. Each condition shows real-time fluorescence detection curves and signal-to-noise ratio analysis (n=3, data presented as mean ± SEM; significant differences determined using GraphPad Prism 7 with Kruskal-Wallis test followed by Dunn’s multiple comparisons test, indicated by different letters, P<0.05), as well as visual observation under 470 nm blue light illumination. Positive samples (+, with plasmid target) and negative controls (-, without plasmid target) were included to evaluate effective signal activation and background fluorescence. The same below
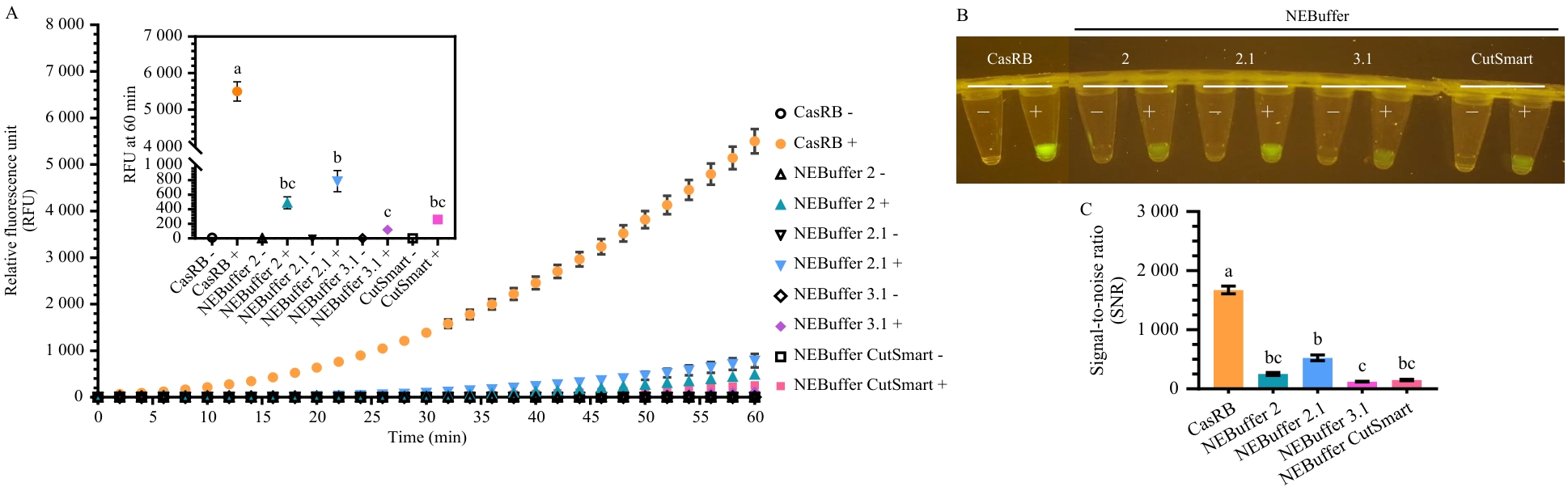
Fig. 3 Performance comparison between CasRB and commercial buffers in the CRISPR/Cas12a fluorescence detection systemA: Real-time fluorescence detection curves; B: Visual observation under 470 nm blue light illumi nation; C: Signal-to-noise ratio of fluorescence detection
| 缓冲液体系Buffer | 信噪比(均值±标准差) Signal-to-noise ratio (Mean ± SD) | 相对信噪比 Relative SNR (CasRB=1) | 单反应成本估算(分) Estimated cost per reaction (CNY cent) | 相对成本 Relative cost (CasRB=1) |
|---|---|---|---|---|
| CasRB | 1 672.8±110.9 | 1.00 | 0.015a | 1 |
| NEBuffer 2 | 253.3±43.6 | 0.15 | 24.54b | 1 636 |
| NEBuffer 2.1 | 525.4±82.8 | 0.31 | 24.54b | 1 636 |
| NEBuffer 3.1 | 122.0±11.4 | 0.07 | 24.54b | 1 636 |
| NEBuffer CutSmart | 149.9±18.2 | 0.09 | 24.54b | 1 636 |
Table 2 Comparison of signal-to-noise ratio (SNR) and estimated cost between CasRB and commercial buffers at the 60 min endpoint
| 缓冲液体系Buffer | 信噪比(均值±标准差) Signal-to-noise ratio (Mean ± SD) | 相对信噪比 Relative SNR (CasRB=1) | 单反应成本估算(分) Estimated cost per reaction (CNY cent) | 相对成本 Relative cost (CasRB=1) |
|---|---|---|---|---|
| CasRB | 1 672.8±110.9 | 1.00 | 0.015a | 1 |
| NEBuffer 2 | 253.3±43.6 | 0.15 | 24.54b | 1 636 |
| NEBuffer 2.1 | 525.4±82.8 | 0.31 | 24.54b | 1 636 |
| NEBuffer 3.1 | 122.0±11.4 | 0.07 | 24.54b | 1 636 |
| NEBuffer CutSmart | 149.9±18.2 | 0.09 | 24.54b | 1 636 |
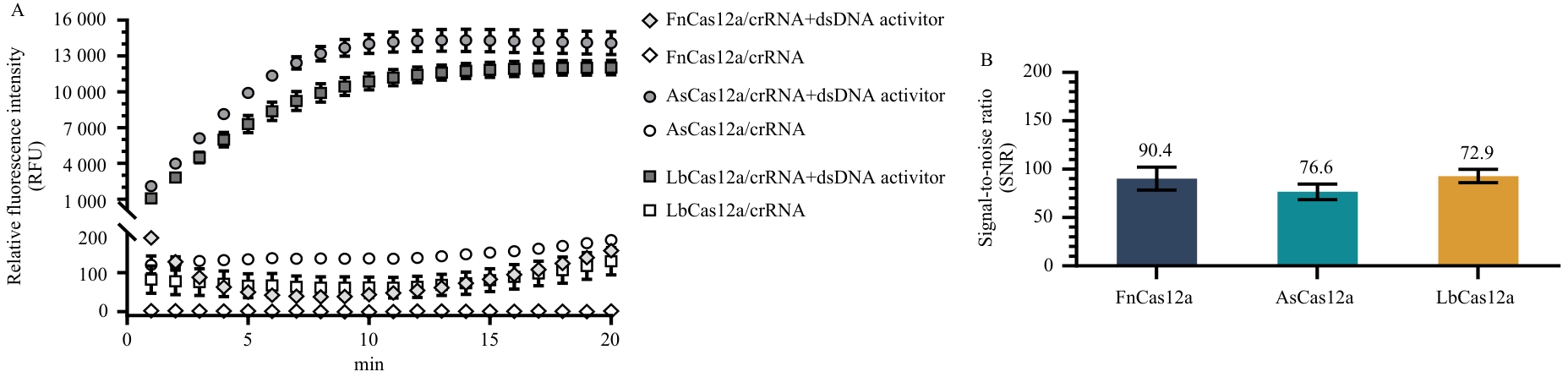
Fig. 4 Evaluation of the applicability of CasRB to different Cas12a enzymesA: Real-time fluorescence detection curves; B: Signal-to-noise ratio of fluorescence detection at 20 min.
| [1] | Chen JS, Ma E, Harrington LB, et al. CRISPR-Cas12a target binding unleashes indiscriminate single-stranded DNase activity [J]. Science, 2018, 360(6387): 436-439. |
| [2] | Xin XL, Su J, Cui HR, et al. Recent advances in clustered regularly interspaced short palindromic repeats/CRISPR-associated proteins system-based biosensors [J]. Biosensors, 2025, 15(3): 155. |
| [3] | Pan LY, Jiang WW, Deng F, et al. Signal-amplification strategy and advances in electrochemistry-based CRISPR/Cas12 biosensing [J]. Chem Eng J, 2025, 508: 161110. |
| [4] | Swarts DC, Jinek M. Mechanistic insights into the cis- and trans-acting DNase activities of Cas12a [J]. Mol Cell, 2019, 73(3): 589-600.e4. |
| [5] | Son H, Park J, Choi YH, et al. Exploring the dynamic nature of divalent metal ions involved in DNA cleavage by CRISPR-Cas12a [J]. Chem Commun, 2022, 58(12): 1978-1981. |
| [6] | Song Y, Xu YW, Wang RR, et al. CRISPR-Cas12a-based nanoparticle biosensor for detection of pathogenic bacteria in food [J]. Microchem J, 2024, 207: 111813. |
| [7] | Ma YJ, Wei HJ, Wang YX, et al. Efficient magnetic enrichment cascade single-step RPA-CRISPR/Cas12a assay for rapid and ultrasensitive detection of Staphylococcus aureus in food samples [J]. J Hazard Mater, 2024, 465: 133494. |
| [8] | 田道明, 周子萱, 杨迎澳, 等. CRISPR-Cas12a介导的适配体荧光传感器快速检测金黄色葡萄球菌 [J]. 中国医药科学, 2024(21): 139-143. |
| Tian DM, Zhou ZX, Yang YA, et al. Rapid detection of Staphylococcus aureus by aptamer fluorescence sensor mediated by CRISPR-Cas12a [J]. China Medicine and Pharmacy, 2024(21): 139-143. | |
| [9] | He W, Liao K, Li RX, et al. Development of a CRISPR/Cas12a-based fluorescent detection method of senecavirus A [J]. BMC Vet Res, 2024, 20(1): 258. |
| [10] | Cao ZZ, Li Z, Jin KK, et al. A rapid and visual detection method for porcine delta coronavirus with single-copy sensitivity based on the CRISPR/Cas12a assay [J]. J Integr Agric, 2025 |
| [11] | 热则古丽·艾科拜尔, 张亚平, 刘浩然, 等. 基于RAA-CRISPR/Cas12a建立牛病毒性腹泻病毒的可视化快速检测方法及其初步应用 [J]. 中国兽医科学, 2025(3): 321-329. |
| Reze Guli A, Zhang YP, Liu HR, et al. Establishment of a visual rapid detection method for bovine viral diarrhea virus based on RAA-CRISPR/Cas12a and its preliminary application [J]. Chinese Veterinary Science, 2025(3): 321-329. | |
| [12] | Shashank PR, Parker BM, Rananaware SR, et al. CRISPR-based diagnostics detects invasive insect pests [J]. Mol Ecol Resour, 2024, 24(1): e13881. |
| [13] | Mu KQ, Ren XJ, Yang H, et al. CRISPR-Cas12a-based diagnostics of wheat fungal diseases [J]. J Agric Food Chem, 2022, 70(23): 7240-7247. |
| [14] | Fang YX, Liu LJ, Zhao WY, et al. Rapid and sensitive detection of Verticillium dahliae from soil using LAMP-CRISPR/Cas12a technology [J]. Int J Mol Sci, 2024, 25(10): 5185. |
| [15] | Guo YF, Tan JJ, Jiao BB, et al. Establishment of a rapid detection system for Phytophthora syringae based on RPA/CRISPR-Cas12a [J]. Crop Prot, 2025, 190: 107106. |
| [16] | Nguyen LT, Rananaware SR, Pizzano BLM, et al. Clinical validation of engineered CRISPR/Cas12a for rapid SARS-CoV-2 detection [J]. Commun Med, 2022, 2: 7. |
| [17] | Dong JJ, Wang SJ, Xu WX, et al. Understanding the stability landscape of LbCas12a by deep analysis of stabilizing mutations and mutation combinations [J]. Protein Sci, 2025, 34(9): e70280. |
| [18] | Tong XH, Li TL, Zhang K, et al. Structure-guided design of Cas12a variants improves detection of nucleic acids [J]. Cell Insight, 2025, 4(2): 100228. |
| [19] | Hwang I, Song YH, Lee S. Enhanced trans-cleavage activity using CRISPR-Cas12a variant designed to reduce steric inhibition by cis-cleavage products [J]. Biosens Bioelectron, 2025, 267: 116859. |
| [20] | Nguyen LT, Smith BM, Jain PK. Enhancement of trans-cleavage activity of Cas12a with engineered crRNA enables amplified nucleic acid detection [J]. Nat Commun, 2020, 11: 4906. |
| [21] | Li YY, University CM, Hu QF, et al. CrRNA conformation-engineered CRISPR-Cas12a system for robust and ultrasensitive nucleic acid detection [J]. Anal Chem, 2025, 97(6): 3617-3624. |
| [22] | Cheng ZH, Luo XY, Yu SS, et al. Tunable control of Cas12 activity promotes universal and fast one-pot nucleic acid detection [J]. Nat Commun, 2025, 16: 1166. |
| [23] | Son H, Park J, Hwang I, et al. Mg2+-dependent conformational rearrangements of CRISPR-Cas12a R-loop complex are mandatory for complete double-stranded DNA cleavage [J]. Proc Natl Acad Sci U S A, 2021, 118(49): e2113747118. |
| [24] | Wang D, Ma DJ, Han JX, et al. CRISPR RNA array-guided multisite cleavage for gene disruption by Cas9 and Cpf1 [J]. ChemBioChem, 2018, 19(20): 2195-2205. |
| [25] | Su F, Wang S, Gao W, et al. Visual detection of multiple Fusarium species via RAA-CRISPR/Cas12a dual fluorescence-lateral flow assay [J]. J Future Foods, 2025 |
| [26] | Chen JL, Huang YE, Xiao B, et al. Development of a RPA-CRISPR-Cas12a assay for rapid, simple, and sensitive detection of Mycoplasma hominis [J]. Front Microbiol, 2022, 13: 842415. |
| [27] | Pan JF, Deng F, Liu Z, et al. Construction of molecular logic gates using heavy metal ions as inputs based on catalytic hairpin assembly and CRISPR-Cas12a [J]. Talanta, 2023, 255: 124210. |
| [28] | Nguyen GT, Schelling MA, Raju A, et al. CRISPR-Cas12a exhibits metal-dependent specificity switching [J]. Nucleic Acids Res, 2024, 52(16): 9343-9359. |
| [29] | Zhu FX, Yu H, Zhao Q. CRISPR/Cas12a-amplified aptamer switch microplate assay for small molecules [J]. Anal Chem, 2024, 96(17): 6853-6859. |
| [30] | Zhang YD, He BS, Zhang YR, et al. An aptasensor based on CRISPR/cas12a-mediated and Mn2+-assisted DNA motor cascade signalling amplification strategy for the detection of T-2 toxin [J]. Sens Actuat B Chem, 2025, 426: 137049. |
| [31] | Liu Y, Liu XY, Wei DY, et al. CoHIT: a one-pot ultrasensitive ERA-CRISPR system for detecting multiple same-site indels [J]. Nat Commun, 2024, 15: 5014. |
| [1] | HUO Guan-zhong, ZHANG Xin-ru, TIAN Shi-jun, LI Jun. Current Progress and Applications of CRISPR/Cas12a Gene Editing Technology in Plants [J]. Biotechnology Bulletin, 2025, 41(6): 1-11. |
| [2] | GAO Chang, ZHUANG Tian-chi, LI Ning, LIU Yun, GU Peng-fei, ZHAO Xin-yi, JI Ming-hui. Gravity-driven Microfluidic Chip Based on RPA-CRISPR/Cas12a for the Rapid Detection of Mycobacterium tuberculosis [J]. Biotechnology Bulletin, 2025, 41(5): 62-69. |
| [3] | TONG De-li, ZHANG Xin, CHEN Jia-qing, HE Hai-sheng. Isolation of Endophytic Bacteria from Blueberry and Its Alleviative Effects on Brassica chinensis L. under Aluminum Stress [J]. Biotechnology Bulletin, 2025, 41(4): 302-311. |
| [4] | ZHAO Chun-duo, LI Yu-e, LIU You-jie, WANG Xin-hang, ZHAO Wei, HUANG Yong-cheng, LI Hu-lin, JI Wen-xiu. Effects of Rotating Cropping and Continuous Cropping on Soil Nutrients, Enzyme Activities and Microbial Community Structure of Rhizosphere Soil in Tobacco [J]. Biotechnology Bulletin, 2025, 41(4): 312-322. |
| [5] | SONG Fen-fen, DUAN Yan-xue, SANG Yu, WANG Ji-peng, PENG Rui, SUN Nian-xi, LI Yong. Characteristics of the Mycosphere Microbial Community in Diseased and Healthy Morchella spp.Soil [J]. Biotechnology Bulletin, 2025, 41(4): 323-334. |
| [6] | XIANG Chun-fan, LI Le-song, WANG Juan, LIANG Yan-li, YANG Sheng-chao, LI Meng-fei, ZHAO Yan. Functional Identification and Expression Analysis of Cinnamonyl Alcohol Dehydrogenase AsCAD in Angelica sinensis [J]. Biotechnology Bulletin, 2025, 41(2): 295-308. |
| [7] | CHEN Mo-yan, ZHU Cheng. Mechanism Study and Application of CRISPR/Cas12a-based Biosensing Platform [J]. Biotechnology Bulletin, 2024, 40(7): 90-98. |
| [8] | JIANG Wen-ping, RAN Qiu-ping, LIU Jia-shu, ZHANG Hui-min, ZHANG Di, JIANG Zheng-bing, LI Hua-nan. Effects of Carbohydrate-binding Modules on the Enzymatic Properties of Xylanase [J]. Biotechnology Bulletin, 2024, 40(5): 269-279. |
| [9] | PAN Ping-ping, XU Zhi-hao, ZHANG Yi-wen, LI Qing, WANG Zhong-hua. Prokaryotic Expression, Subcellular Localization and Expression Analysis of PcCHS Gene from Polygonatum cyrtonema Hua [J]. Biotechnology Bulletin, 2024, 40(5): 280-289. |
| [10] | LI Xue, LI Rong-ou, KONG Mei-yi, HUANG Lei. The Growth Promoting Effect of Bacillus amyloliquefaciens SQ-2 on Rice [J]. Biotechnology Bulletin, 2024, 40(2): 109-119. |
| [11] | HU Hai-yang, YING Wan-qin, HE Jun, LV Zhi-xian, XIE Xiao-ping, DENG Zhong-liang. Establishment and Application of ERA Real-time Fluorescence Method for Rapid Detection of Mycoplasma pneumoniae [J]. Biotechnology Bulletin, 2022, 38(9): 264-270. |
| [12] | ZHAO Zhong-juan, YANG Kai, HU Jin-dong, WEI Yan-li, LI Ling, XU Wei-sheng, LI Ji-shun. Effects of Trichoderma harzianum ST02 on the Growth of Peppermint and Physicochemical Properties of Root Zone Soil Under Salt Stress [J]. Biotechnology Bulletin, 2022, 38(7): 224-235. |
| [13] | YUAN Cun-xia, LI Yan-nan, ZHANG Xiao-chong, YANG Rui, LIU Jian-li, LI Jing-yu. Physiological and Biochemical Response Characteristics of Bacillus sp. ZJS3 Under As3+ Stress [J]. Biotechnology Bulletin, 2022, 38(7): 236-246. |
| [14] | WANG Xiao-qin, HUANG Yin-ping, WANG Wei-qian, WU Ping, QUAN Shu. Expression and Purification of the MLL3SET Protein with a Site-directed Mutation of an Unnatural Amino Acid [J]. Biotechnology Bulletin, 2022, 38(3): 194-202. |
| [15] | JIA Hai-hong, LI Bing-qing. Research Progress in the Post-translational Modification of Superoxide Dismutase [J]. Biotechnology Bulletin, 2022, 38(2): 237-244. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||