








Biotechnology Bulletin ›› 2025, Vol. 41 ›› Issue (5): 62-69.doi: 10.13560/j.cnki.biotech.bull.1985.2024-0955
Previous Articles Next Articles
GAO Chang1( ), ZHUANG Tian-chi2, LI Ning3, LIU Yun4, GU Peng-fei1, ZHAO Xin-yi5, JI Ming-hui2(
), ZHUANG Tian-chi2, LI Ning3, LIU Yun4, GU Peng-fei1, ZHAO Xin-yi5, JI Ming-hui2( )
)
Received:2024-09-30
Online:2025-05-26
Published:2025-06-05
Contact:
JI Ming-hui
E-mail:1975480691@qq.com;jiminghui@njmu.edu.cn
GAO Chang, ZHUANG Tian-chi, LI Ning, LIU Yun, GU Peng-fei, ZHAO Xin-yi, JI Ming-hui. Gravity-driven Microfluidic Chip Based on RPA-CRISPR/Cas12a for the Rapid Detection of Mycobacterium tuberculosis[J]. Biotechnology Bulletin, 2025, 41(5): 62-69.
| 名称 Primer | 序列 Primer sequence (5′-3′) | 长度 Length (bp) |
|---|---|---|
| F1 | GTCATGTCAGGTGGTTCATCGAGGAGGTACC | 31 |
| R1 | GCCCACTCCGAATCGTGCTGACCGCGGATCT | 31 |
| F2 | GTCATGTCAGGTGGTTCATCGAGGAGGTACC | 31 |
| R2 | TGCCCACTCCGAATCGTGCTGACCGCGGATC | 31 |
| F3 | GGTCATGTCAGGTGGTTCATCGAGGAGGTAC | 31 |
| R3 | TGCCCACTCCGAATCGTGCTGACCGCGGATC | 31 |
| F4 | GTCATGTCAGGTGGTTCATCGAGGAGGTACC | 31 |
| R4 | CCCACTCCGAATCGTGCTGACCGCGGATCTC | 31 |
| crRNA-1 | GATGAGCCCGCGGCCTATCAGCTTG | 25 |
| crRNA-2 | TGGTGGGGTGACGGCCTACCAAGGC | 25 |
| crRNA-3 | ACGACGGGTAGCCGGCCTGAGAGG | 24 |
| crRNA-4 | GGAGGTGGAGAAGAAGCACCGGCC | 24 |
| ssDNA报告探针 | 6-FAM-TTATT-BHQ-1 | 5 |
Table 1 Primers, crRNA and ssDNA reporter used in this study
| 名称 Primer | 序列 Primer sequence (5′-3′) | 长度 Length (bp) |
|---|---|---|
| F1 | GTCATGTCAGGTGGTTCATCGAGGAGGTACC | 31 |
| R1 | GCCCACTCCGAATCGTGCTGACCGCGGATCT | 31 |
| F2 | GTCATGTCAGGTGGTTCATCGAGGAGGTACC | 31 |
| R2 | TGCCCACTCCGAATCGTGCTGACCGCGGATC | 31 |
| F3 | GGTCATGTCAGGTGGTTCATCGAGGAGGTAC | 31 |
| R3 | TGCCCACTCCGAATCGTGCTGACCGCGGATC | 31 |
| F4 | GTCATGTCAGGTGGTTCATCGAGGAGGTACC | 31 |
| R4 | CCCACTCCGAATCGTGCTGACCGCGGATCTC | 31 |
| crRNA-1 | GATGAGCCCGCGGCCTATCAGCTTG | 25 |
| crRNA-2 | TGGTGGGGTGACGGCCTACCAAGGC | 25 |
| crRNA-3 | ACGACGGGTAGCCGGCCTGAGAGG | 24 |
| crRNA-4 | GGAGGTGGAGAAGAAGCACCGGCC | 24 |
| ssDNA报告探针 | 6-FAM-TTATT-BHQ-1 | 5 |

Fig. 1 Screening of RPA primers and crRNA targeting the IS6110 gene of M. tuberculosisA: Detection of RPA amplification products from different primers by agarose gel electrophoresis. B: Detection of RPA amplification specificity by agarose gel electrophoresis (1: Mycobacterium tuberculosis; 2: Staphylococcus aureus; 3: Pseudomonas aeruginosa; 4: Klebsiella pneumoniae. C: Fluorescence intensity of CRISPR/Cas12a reactions with different crRNAs. Each test was repeated three times, and fluorescence intensity is shown in the figure as mean±SD (NTC: negative control, ddH2O; * P<0.05, ** P<0.01, **** P<0.000 1. The same below)
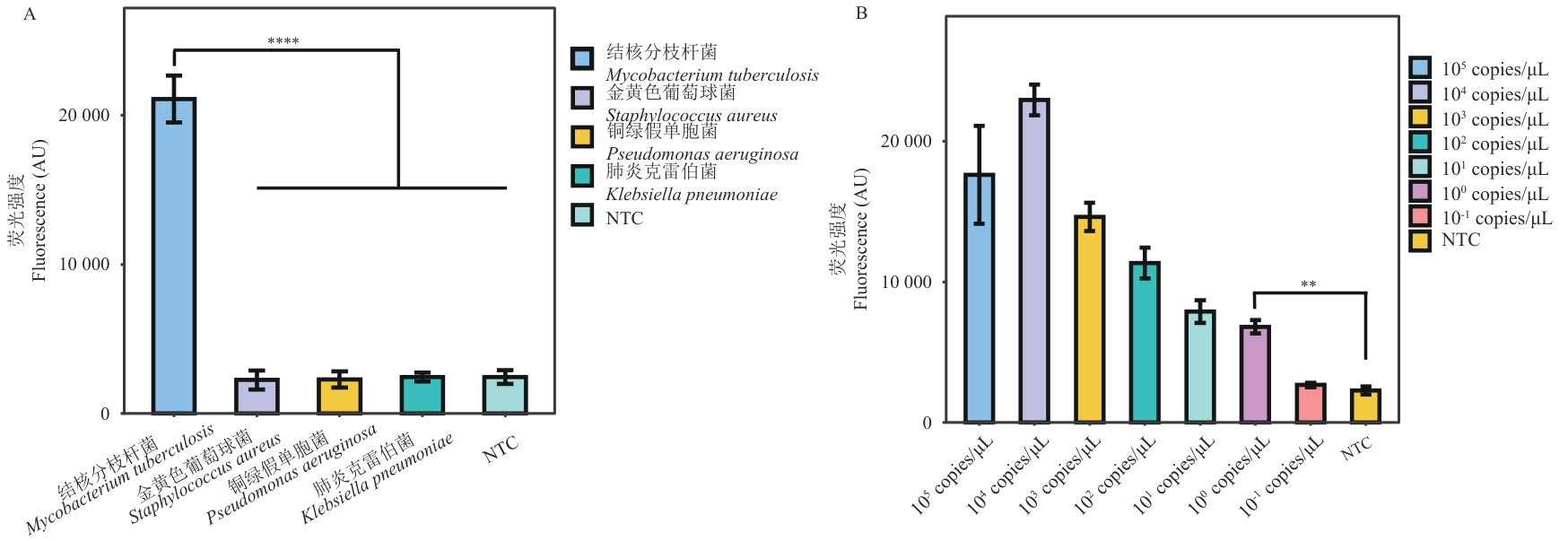
Fig. 2 RPA-CRISPR/Cas12a system testingA: Specificity assessment of RPA-CRISPR/Cas12a targeting the IS6110 gene of M. tuberculosis. B: Sensitivity assessment of RPA-CRISPR/Cas12a targeting the IS6110 gene of M. tuberculosis. Each test was repeated three times, and fluorescence intensity is shown in the figure as mean±SD
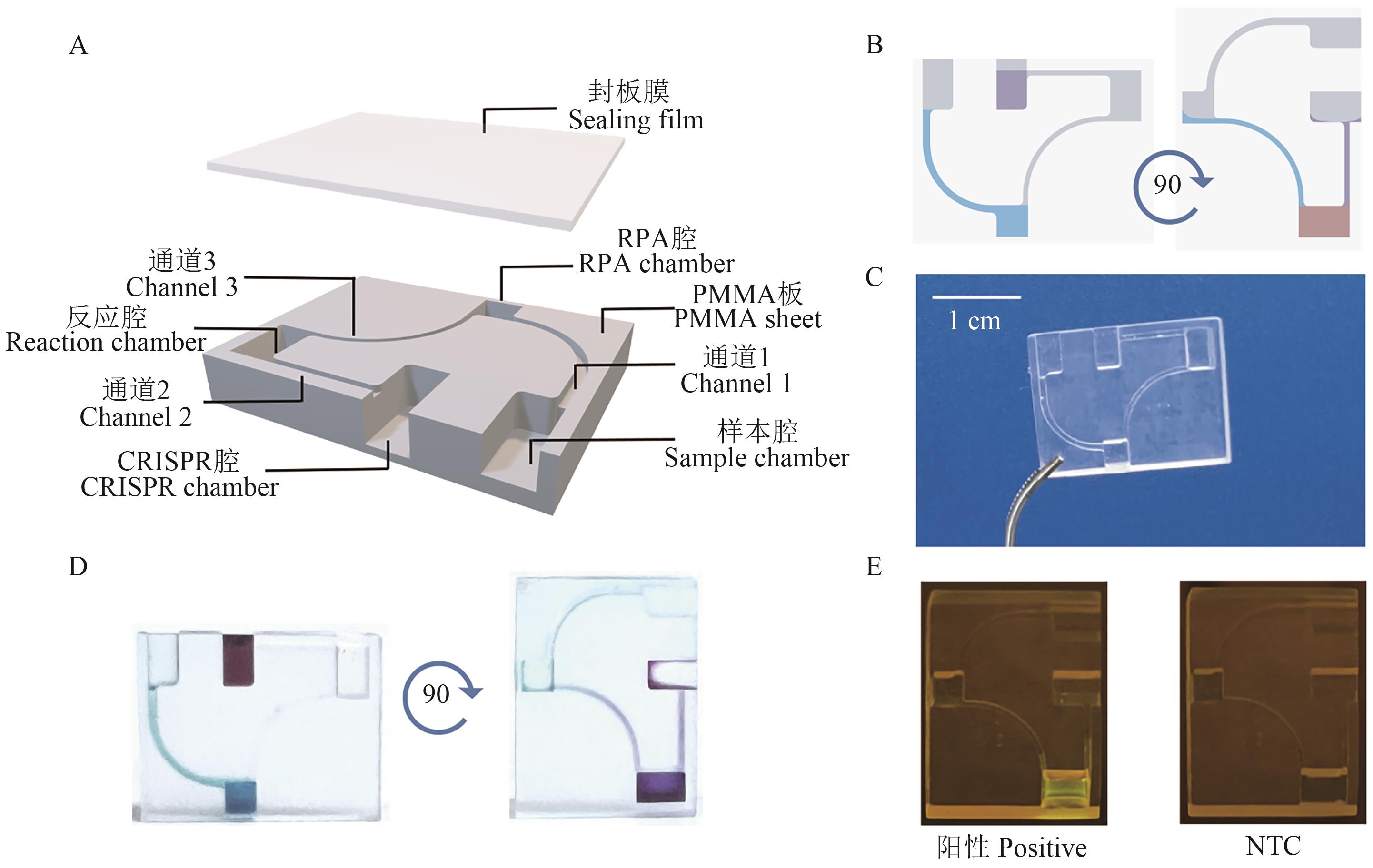
Fig. 3 Gravity-driven microfluidic chip for RPA-CRISPR/Cas12a detectionA: Schematic diagram of the chip structure. B: Schematic diagram of the chip working principle. C: Image of the chip. D: Chip testing image using dye. E: Detection results of RPA-CRISPR/Cas12a assay on the chip
RPA-CRISPR/Cas12a片上检测 RPA-CRISPR/Cas12a for on-chip detection | 痰培养 Sputum culture | 灵敏度 Sensitivity (%) | 特异性 Specificity (%) | 阳性预测值 Positive predictive value (%) | 阴性预测值 Negative predictive value (%) | 准确性 Accuracy (%) | Kappa值 | ||
|---|---|---|---|---|---|---|---|---|---|
阳性 Positive | 阴性 Negative | 合计 Total | |||||||
| 阳性 Positive | 123 | 9 | 132 | 91.11 | 94.34 | 93.19 | 92.59 | 92.86 | 0.856 |
| 阴性 Negative | 12 | 150 | 162 | ||||||
| 合计 Total | 135 | 159 | 294 | ||||||
Table 2 Detection of M. tuberculosis in sputum samples using RPA-CRISPR/Cas12a and sputum culture methods
RPA-CRISPR/Cas12a片上检测 RPA-CRISPR/Cas12a for on-chip detection | 痰培养 Sputum culture | 灵敏度 Sensitivity (%) | 特异性 Specificity (%) | 阳性预测值 Positive predictive value (%) | 阴性预测值 Negative predictive value (%) | 准确性 Accuracy (%) | Kappa值 | ||
|---|---|---|---|---|---|---|---|---|---|
阳性 Positive | 阴性 Negative | 合计 Total | |||||||
| 阳性 Positive | 123 | 9 | 132 | 91.11 | 94.34 | 93.19 | 92.59 | 92.86 | 0.856 |
| 阴性 Negative | 12 | 150 | 162 | ||||||
| 合计 Total | 135 | 159 | 294 | ||||||
| 1 | Xiao J, Li JQ, Quan ST, et al. Development and preliminary assessment of a CRISPR-Cas12a-based multiplex detection of Mycobacterium tuberculosis complex [J]. Front Bioeng Biotechnol, 2023, 11: 1233353. |
| 2 | World Health O. Global tuberculosis report 2021 [R]. World Health Organization, 2021. |
| 3 | World Health O. Global tuberculosis report 2023 [R]. World Health Organization, 2023. |
| 4 | Zhang D, Yu F, Han DS, et al. ddPCR provides a sensitive test compared with GeneXpert MTB/RIF and mNGS for suspected Mycobacterium tuberculosis infection [J]. Front Cell Infect Microbiol, 2023, 13: 1216339. |
| 5 | Chen YJ, Zhu YY, Wang XF, et al. Gravity-driven and rotation-controlled microfluidic chip for point-of-care nucleic acid detection in the fully closed environment [J]. Talanta, 2024, 267: 125258. |
| 6 | Mosquera-Restrepo SF, Zuberogoïtia S, Gouxette L, et al. A Mycobacterium tuberculosis fingerprint in human breath allows tuberculosis detection [J]. Nat Commun, 2022, 13(1): 7751. |
| 7 | Maréchal A, Zou LE. RPA-coated single-stranded DNA as a platform for post-translational modifications in the DNA damage response [J]. Cell Res, 2015, 25(1): 9-23. |
| 8 | Pike AM, Friend CM, Bell SP. Distinct RPA functions promote eukaryotic DNA replication initiation and elongation [J]. Nucleic Acids Res, 2023, 51(19): 10506-10518. |
| 9 | Hefel A, Honda M, Cronin N, et al. RPA complexes in Caenorhabditis elegans meiosis; unique roles in replication, meiotic recombination and apoptosis [J]. Nucleic Acids Res, 2021, 49(4): 2005-2026. |
| 10 | Nguyen LT, Gurijala J, Rananaware SR, et al. CRISPR-ENHANCE: an enhanced nucleic acid detection platform using Cas12a [J]. Methods, 2022, 203: 116-124. |
| 11 | Qiu XT, Xu S, Liu XP, et al. CRISPR/Cas12a-based diagnostic platform accurately detects Nocardia farcinica targeting a novel species-specific gene [J]. Front Cell Infect Microbiol, 2022, 12: 884411. |
| 12 | Phuphisut O, Poodeepiyasawat A, Yoonuan T, et al. Ov-RPA-CRISPR/Cas12a assay for the detection of Opisthorchis viverrini infection in field-collected human feces [J]. Parasit Vectors, 2024, 17(1): 80. |
| 13 | 徐银, 刘婷, 徐慧, 等. PCR-CRISPR/Cas12a华支睾吸虫囊蚴检测方法的建立和应用 [J]. 中国寄生虫学与寄生虫病杂志, 2023, 41(4): 421-426, 433. |
| Xu Y, Liu T, Xu H, et al. Establishment and application of PCR-CRISPR/Cas12a-based detection method for Clonorchis sinensis metacercaria [J]. Chin J Parasitol Parasit Dis, 2023, 41(4): 421-426, 433. | |
| 14 | Wang T, Bai LL, Wang GL, et al. SATCAS: a CRISPR/Cas13a-based simultaneous amplification and testing platform for one-pot RNA detection and SNPs distinguish in clinical diagnosis [J]. Biosens Bioelectron, 2024, 263: 116636. |
| 15 | 侯雅超, 邢微微, 王亚楠, 等. 针对副溶血性弧菌toxR基因的RPA-CRISPR/Cas13a荧光快速检测方法的建立及应用 [J]. 中华医院感染学杂志, 2024, 34(14): 2081-2086. |
| Hou YC, Xing WW, Wang YN, et al. Establishment and application of RPA-CRISPR/Cas13a fluorescence rapid detection method for toxR gene of Vibrio parahaemolyticus [J]. China Ind Econ, 2024, 34(14): 2081-2086. | |
| 16 | 王文涛, 赵良, 侯立功, 等. RPA-CRISPR/Cas12a在病原微生物检测中的研究进展 [J]. 微生物学通报, 2024, 51(8): 2785-2796. |
| Wang WT, Zhao L, Hou LG, et al. Research progress of RPA-CRISPR/Cas12a in the detection of pathogenic microorganisms [J]. Microbiol China, 2024, 51(8): 2785-2796. | |
| 17 | 张徐俞, 黄俊, 杨稳, 等. 重组酶聚合酶扩增结合CRISPR-Cas12a快速检测十足目虹彩病毒1方法的建立 [J]. 微生物学通报, 2021, 48(12): 4980-4988. |
| Zhang XY, Huang J, Yang W, et al. Rapid detection of decapod iridescent virus 1 by recombinase polymerase amplification combined with CRISPR-Cas12a [J]. Microbiol China, 2021, 48(12): 4980-4988. | |
| 18 | Sun YY, Yu L, Liu CX, et al. One-tube SARS-CoV-2 detection platform based on RT-RPA and CRISPR/Cas12a [J]. J Transl Med, 2021, 19(1): 74. |
| 19 | Zhuang TC, Gao C, Zhao WW, et al. A minimal transcription template-based amplification-free CRISPR-Cas13a strategy for DNA detection [J]. Biosens Bioelectron, 2025, 270: 116918. |
| 20 | Hu ML, Qiu ZQ, Bi ZR, et al. Photocontrolled crRNA activation enables robust CRISPR-Cas12a diagnostics [J]. Proc Natl Acad Sci USA, 2022, 119(26): e2202034119. |
| 21 | Zhang WS, Pan JB, Li F, et al. Reverse transcription recombinase polymerase amplification coupled with CRISPR-Cas12a for facile and highly sensitive colorimetric SARS-CoV-2 detection [J]. Anal Chem, 2021, 93(8): 4126-4133. |
| 22 | Guo YF, Xia HM, Dai TT, et al. RPA-CRISPR/Cas12a mediated isothermal amplification for visual detection of Phytophthora sojae [J]. Front Cell Infect Microbiol, 2023, 13: 1208837. |
| 23 | Hajian R, Balderston S, Tran T, et al. Detection of unamplified target genes via CRISPR-Cas9 immobilized on a graphene field-effect transistor [J]. Nat Biomed Eng, 2019, 3(6): 427-437. |
| 24 | 林正龙, 张蓓英. 基因芯片技术快速检测结核分枝杆菌耐药基因的应用价值研究 [J]. 医学理论与实践, 2024, 37(17): 2893-2896. |
| Lin ZL, Zhang BY. Evaluation of gene chips for the rapid diagnosis of drug-resistance genes in Mycobacterium tuberculosis [J]. J Med Theory Pract, 2024, 37(17): 2893-2896. | |
| 25 | 陈墨岩, 祝诚. 基于CRISPR/Cas12a的生物传感平台的机制研究及应用 [J]. 生物技术通报, 2024, 40(7): 90-98. |
| Chen MY, Zhu C. Mechanism study and application of CRISPR/Cas12a-based biosensing platform [J]. Biotechnol Bull, 2024, 40(7): 90-98. | |
| 26 | Bai YM, Ji JC, Ji FD, et al. Recombinase polymerase amplification integrated with microfluidics for nucleic acid testing at point of care [J]. Talanta, 2022, 240: 123209. |
| 27 | Xia LP, Yin JX, Zhuang JJ, et al. Adsorption-free self-priming direct digital dual-crRNA CRISPR/Cas12a-assisted chip for ultrasensitive detection of pathogens [J]. Anal Chem, 2023, 95(10): 4744-4752. |
| 28 | Khan MS, Ning Y, Jinou C, et al. Are global tuberculosis control targets overlooking an essential indicator? Prolonged delays to diagnosis despite high case detection rates in Yunnan, China [J]. Health Policy Plan, 2017, 32(): i15-i21. |
| 29 | Liu Y, Chao Z, Ding W, et al. Correction: a multiplex RPA-CRISPR/Cas12a-based POCT technique and its application in human papillomavirus (HPV) typing assay [J]. Cell Mol Biol Lett, 2024, 29(1): 49. |
| 30 | Fozouni P, Son S, de León Derby MD, et al. Amplification-free detection of SARS-CoV-2 with CRISPR-Cas13a and mobile phone microscopy [J]. Cell, 2021, 184(2): 323-333.e9. |
| 31 | Zhang XY, Tian Y, Xu L, et al. CRISPR/Cas13-assisted hepatitis B virus covalently closed circular DNA detection [J]. Hepatol Int, 2022, 16(2): 306-315. |
| 32 | Cheng M, Tan CW, Xiang B, et al. Chain hybridization-based CRISPR-lateral flow assay enables accurate gene visual detection [J]. Anal Chim Acta, 2023, 1270: 341437. |
| [1] | CHEN Mo-yan, ZHU Cheng. Mechanism Study and Application of CRISPR/Cas12a-based Biosensing Platform [J]. Biotechnology Bulletin, 2024, 40(7): 90-98. |
| [2] | WANG Ling-ling, MA Ai-hong, SONG Qian-jin, WANG Xiao-long, CAO Xiao-zhen, LIU Zhi-feng, CHEN Ya-fei, LI Ji-dong. Establishment and Application of Dual RPA-LFD Detection Method for Capripox Virus and Orf Virus [J]. Biotechnology Bulletin, 2024, 40(5): 103-111. |
| [3] | CHEN Xiao-lin, LIU Yang-er, XU Wen-tao, GUO Ming-zhang, LIU Hui-lin. Application of Synthetic Biology Based Whole-cell Biosensor Technology in the Rapid Detection of Food Safety [J]. Biotechnology Bulletin, 2023, 39(1): 137-149. |
| [4] | WEI Qian, LIU Xiao-ning, ZHAO Jie. FoxAl Regulating CYP6B6 Expression Under 2-tridecanone Stress in Helicoverpa armigera [J]. Biotechnology Bulletin, 2022, 38(5): 84-92. |
| [5] | FU Zhi-qiang, XIONG Yan. Research Progress on Portable Bio-optical Sensors [J]. Biotechnology Bulletin, 2021, 37(3): 219-226. |
| [6] | LI Tianjingwei, ZOU Xiao-xiao, ZHU Jun, BAO Shi-xiang. Selection and Validation of Reference Genes for Quantitative Real-time PCR in Caulerpa lentillifera Under Stress Conditions [J]. Biotechnology Bulletin, 2021, 37(10): 266-276. |
| [7] | LI Xin-shen, HUANG Xiao-mei, WU Shu-xiu, HUANG Rui-rong, WEI Lin-gen, HUA Ju-ling. Rapid Detection of Plant Bacterial Wilt by Loop-mediated Isothermal Amplification [J]. Biotechnology Bulletin, 2021, 37(1): 272-281. |
| [8] | ZHAO Ying, WANG Nan, LU An-xiang, FENG Xiao-yuan, GUO Xiao-jun, LUAN Yun-xia. Application in the Detection of Fungal Toxins by Nucleic Acid Aptamer Lateral Flow Chromatography Analysis Technique [J]. Biotechnology Bulletin, 2020, 36(8): 217-227. |
| [9] | GAO Wei-fang, ZHANG Li-ping, ZHU Peng. Recent Progress on Isothermal Amplification Technology and Its Combination with CRISPR in Rapid Detection of Microorganisms [J]. Biotechnology Bulletin, 2020, 36(5): 22-31. |
| [10] | LV Ji-zhou, WU Shao-qiang, ZHANG Zhou, DENG Jun-hua, YUAN Xiang-fen, WANG Cai-xia, FENG Chun-yan, LIN Xiang-mei. Development of a Real-time Fluorescent Double Reverse-Transcription Recombinase Polymerase Amplification Method and Its Application in Detecting SARS-CoV-2 in Food [J]. Biotechnology Bulletin, 2020, 36(11): 238-244. |
| [11] | WANG Qi, YAN Chun-lei, GAO Hong-wei, WU Wei, YANG Qing-li. Research Progress of DNA Aptasensors for Foodborne Pathogen Detection [J]. Biotechnology Bulletin, 2020, 36(11): 245-258. |
| [12] | LIN Hui-jiao, YANG Hua-wei, GU Heng-sen, JIANG Xiang, ZHANG Hai-lei, LIU Yu-chen, ZHOU Er-xun. Development of Dipstick for the Rapid Detection of Three Important Monilinia Species on Fruits [J]. Biotechnology Bulletin, 2019, 35(6): 205-212. |
| [13] | GUO Pei, ZHAO Long, HU He. A Rapid Method of Detecting Viable Legionella pneumophila in the Water Environment of Public Places [J]. Biotechnology Bulletin, 2019, 35(3): 203-209. |
| [14] | LI Zi-wei, DENG Zhong-liang. Application of a Loop-mediated Isothermal Amplification Method for Rapid Diagnosis of Francisella tularensis [J]. Biotechnology Bulletin, 2019, 35(2): 212-217. |
| [15] | ZHANG Wei, LI Zhi-xin, FU Chun-jiang, LIU Wei-ping. Development of a Colloidal Gold Immunochromatographic Test Strip for the Detection of Potato Virus S [J]. Biotechnology Bulletin, 2019, 35(12): 184-188. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||