








Biotechnology Bulletin ›› 2021, Vol. 37 ›› Issue (11): 257-266.doi: 10.13560/j.cnki.biotech.bull.1985.2020-1508
Previous Articles Next Articles
LIU Shan1,2( ), YE Wei2, ZHU Mu-zi2, LI Sai-ni2, DENG Zhang-shuang1(
), YE Wei2, ZHU Mu-zi2, LI Sai-ni2, DENG Zhang-shuang1( ), ZHANG Wei-min2(
), ZHANG Wei-min2( )
)
Received:2020-12-13
Online:2021-11-26
Published:2021-12-03
Contact:
DENG Zhang-shuang,ZHANG Wei-min
E-mail:shanliu1995@126.com;dzs163@163.com;wmzhang@gdim.cn
LIU Shan, YE Wei, ZHU Mu-zi, LI Sai-ni, DENG Zhang-shuang, ZHANG Wei-min. Cloning,Expression and Characterization of a Novel Acyltransferase GPAT[J]. Biotechnology Bulletin, 2021, 37(11): 257-266.
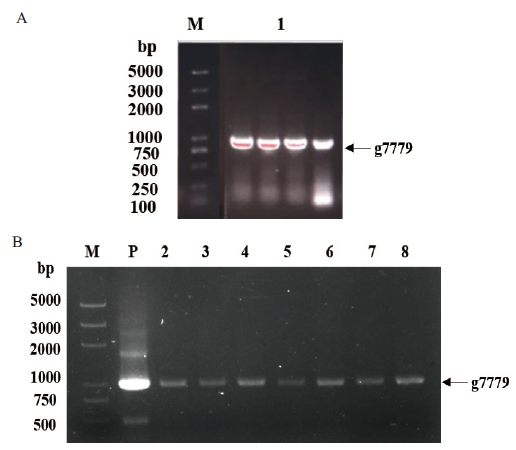
Fig.1 PCR verification of gene g7779 A:Cloning of gene g7779. B:PCR verification of bacterial solution. M:DNA marker 2K Plus. 1:Gene g7779. P:Positive control. 2-8:Positive monoclonal of pET28a-g7779

Fig.2 Purification and Western-Blot analysis of GPAT A:Optimization of the expression condition of GPAT. M:Blue Plus II protein marker. 1, 3, 5, 7, 9 and 11:Supernatant of condition ①, ②, ③, ④, ⑤ and ⑥. 2, 4, 6, 8, 10 and 12:Sediment of condition ①, ②, ③, ④, ⑤ and ⑥. B:Purification of GPAT. M:Blue Plus Ⅱ protein marker. 1:Uninduced total protein. 2:Total protein of supernatant after induction. 3-5:Binding buffer. 6:68 mmol/L imidazole eluent. 7:116 mmol/L imidazole eluent. C:Western-Blot analysis of GPAT. M:EasySee Western marker. 1:Uninduced total protein. 2:Total protein of supernatant after induction. 3:Binding buffer. 4:68 mmol/L imidazole eluent. 5:116 mmol/L imidazole eluent
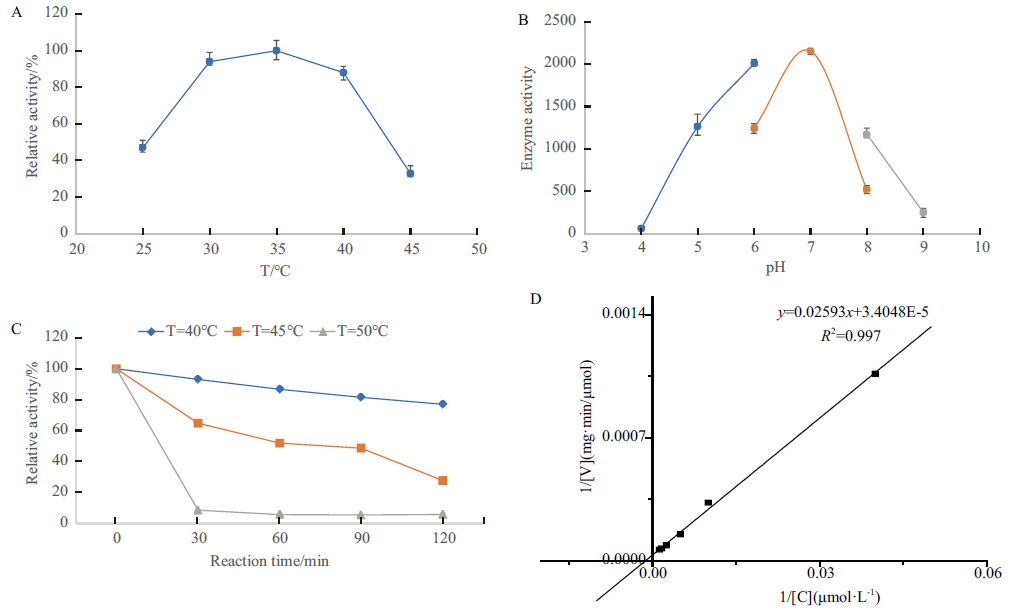
Fig.5 Characterization of enzymatic properties of GPAT A:Optimal reaction temperature of GPAT. B:Optimal reaction pH of GPAT. C:Thermostability of GPAT. D:Enzyme kinetics of GPAT
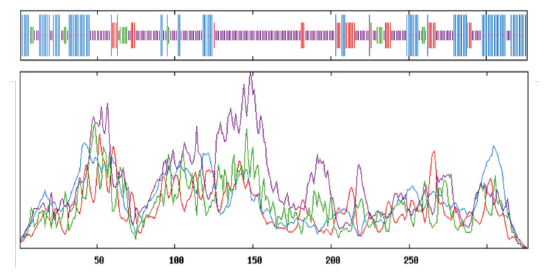
Fig.7 Secondary structure analysis of GPAT The order of vertical lines from long to short are:α - helix(blue),extend strand(red),β - turn(green)and random coil(purple)
| [1] | 范东东, 孙铭娟, 王梁华, 等. 酰基转移酶研究[J]. 生命的化学, 2008, 28(6): 701-703. |
| Fan DD, Sun MJ, Wang LH, et al. Study on Acyltransferase[J]. Chemistry of Life, 2008, 28(6): 701-703. | |
| [2] |
Park YC, Shaffer CEH, Bennett GN. Microbial formation of esters[J]. Applied Microbiology and Biotechnology, 2009, 85(1): 13-25.
doi: 10.1007/s00253-009-2170-x URL |
| [3] | 张利华, 陈献忠, 陈振, 等. 热带假丝酵母肉毒碱乙酰基转移酶基因的删除及功能鉴定[J]. 食品与生物技术学报, 2018, 37(8): 880-887. |
| Zhang LH, Chen XZ, Chen Z, et al. Carnitine acetyltransferase gene disruption and function analysis in Candida Tropicalis[J]. Journal of Food Science and Biotechnology, 2018, 37(8): 880-887. | |
| [4] | 葛文雪, 陈润, 白嘉诚, 等. 结核分枝杆菌硫醇乙酰基转移酶基因敲除株的构建及其生物学特性分析[J]. 微生物与感染, 2019, 14(5): 282-288. |
| Ge WX, Chen R, Bai JC, et al. A mycothiol acetyltransferase knockout in Mycobacterium tuberculosis and its biological characteristics[J]. Journal of Microbes and Infections, 2019, 14(5): 282-288. | |
| [5] | 曹珺. 高山被孢霉中二酰甘油酰基转移酶的筛选鉴定及功能探究[D]. 江南大学, 2019. |
| Cao J. Screening and identification of diacylglycerol acyltransferase from Mortierella alpina and its effect on lipid accumulation[D]. Jiangnan University, 2019. | |
| [6] | 刘雨雨, 莫婷, 王晓晖, 等. 植物来源BAHD酰基转移酶家族研究进展[J]. 中国中药杂志, 2016, 41(12): 2175-2182. |
| Liu YY, Mo T, Wang XH, et al. Research progress of plant BAHD acyltransferase family[J]. China Journal of Chinese Materia Medica, 2016, 41(12): 2175-2182. | |
| [7] |
Xu J, Tan H, Chen Y, et al. Lithocarpins A-D:four tenellone-macrolide conjugated[4 + 2]hetero-adducts from the deep-sea derived fungus Phomopsis lithocarpus FS508[J]. Organic Chemistry Frontiers, 2018, 5: 1792-1797.
doi: 10.1039/C8QO00095F URL |
| [8] | Garvey GS, Mccormick SP, Alexander NJ, et al. Structural and functional characterization of TRI3 trichothecene 15-O-acetyltransferase from Fusarium sporotrichioides[J]. Protein Science, 2009, 18(4): 747-761. |
| [9] |
Dastjerdeh MS, Marashiyan M, Boroujeni MB, et al. In silico analysis of different signal peptides for the secretory production of recombinant human keratinocyte growth factor in Escherichia coli[J]. Computational Biology and Chemistry, 2019, 80: 225-233.
doi: S1476-9271(18)30522-X pmid: 30999249 |
| [10] | Ropón-Palacios G, Chenet-Zuta ME, Otazu K, et al. Novel multi-epitope protein containing conserved epitopes from different Leishmania species as potential vaccine candidate:integrated immunoinformatics and molecular dynamics approach[J]. Computational Biology and Chemistry, 2019, 83: 1-11. |
| [11] | Jamal, Shaheen, Sunita. Study of transmembraneous protein using bioinformatics and data mining[J]. Asian Journal of Bio Science, 2014, 1(9): 71-75. |
| [12] |
Sevindik E. Comparative and phylogenetic analysis of RuBisCO large subunit(rbcL)proteins in some Sideritis L. (Lamiaceae)species:A bioinformatic approach[J]. Genetika-Belgrade, 2019, 51(1): 69-80.
doi: 10.2298/GENSR1901069S |
| [13] | 吴姝, 伊正君, 付玉荣. 结核分枝杆菌Pst S1蛋白结构与功能的生物信息学分析[J]. 中国病原生物学杂志, 2019, 14(7): 806-810+821. |
| Wu S, Yin ZJ, Fu YR. Bioinformatic analysis of the structure and function of Pst S1 from Mycobacterium tuberculosis[J]. Journal of Pathogen Biology, 2019, 14(7): 806-810+821. | |
| [14] | 钱锦, 李丽, 常爱平, 等. 鞘氨醇单胞菌中威兰胶合成关键基因welE和welC的生物信息学分析[J]. 河南师范大学学报:自然科学版, 2020, 48(1): 82-89. |
| Qian J, Li L, Chang AP, et al. Bioinformatics analysis of the key genes welE and welC in the synjournal of Welan gum from Sphingomonas sp.[J]. Journal of Henan Normal University:Natural Science Edition, 2020, 48(1): 82-89. | |
| [15] | 方智振, 姜翠翠, 周丹蓉, 等. 基于转录组的‘三月李’及其红肉突变体ARF基因家族鉴定及分析[J]. 应用与环境生物学报, 2019, 25(6): 1388-1395. |
| Fang ZZ, Jiang CC, Zhou DR, et al. Analysis of the ARF gene family of ‘Sanyueli’ plum(Prunus salicina LindL.)and its red-fleshed mutant based on transcriptome[J]. Chinese Journal of Applied and Environmental Biology, 2019, 25(6): 1388-1395. | |
| [16] |
Qiu JJ, Wang DQ, Ma YF, et al. Identification and characterization of serine acetyltransferase encoded by the Mycobacterium tuberculosis Rv2335 gene[J]. International Journal of Molecular Medicine, 2013, 31(5): 1229-1233.
doi: 10.3892/ijmm.2013.1298 URL |
| [17] | 左泽红, 魏韬, 郭丽琼, 等. 平菇高丝氨酸乙酰基转移酶基因的克隆及异源表达优化[J]. 食品工业科技, 2019, 40(13): 64-70. |
| Zuo ZH, Wei T, Guo LQ, et al. Cloning and heterologous expression optimization of homoserine acetyltransferase gene from Pleurotus ostreatus[J]. Science and Technology of Food Industry, 2019, 40(13): 64-70. | |
| [18] | 钱玉梅, 李红侠, 李秀丽, 等. ‘赤霞珠’葡萄果实中儿茶素没食子酰基化转移酶蛋白的纯化与鉴定[J]. 果树学报, 2020, 37(8): 1132-1143. |
| Qian YM, Li HX, Li XL, et al. Purification and identification of galloyltransferase involved in catechin metabolism in ‘Cabernet Sauvignon’ grape[J]. Journal of Fruit Science, 2020, 37(8): 1132-1143. | |
| [19] | 甄军波, 刘琳琳, 杜海英, 等. 海岛棉GbGPAT2基因的克隆与表达分析[J]. 西南农业学报, 2020, 33(3): 503-508. |
| Zhen JB, Liu LL, Du HY, et al. Cloning and Expression Analysis of GbGPAT2 from Gossypium barbadense[J]. Southwest China Journal of Agricultural Sciences, 2020, 33(3): 503-508. | |
| [20] | 刘小琴, 杨岩, 吴喆瑜, 等. 多点突变提高α-L-鼠李糖苷酶热稳定性[J]. 食品与发酵工业, 2019, 45(6): 23-29. |
| Liu XQ, Yang Y, Wu ZY, et al. Improvement of thermal stability of α-L-rhamnosidase by multiple point mutation[J]. Food and Fermentation Industries, 2019, 45(6): 23-29. | |
| [21] | 刘晓彤, 邬敏辰, 殷欣, 等. 二硫键对提高木聚糖酶AoXyn11A热稳定性的作用[J]. 食品与生物技术学报, 2014, 33(10): 1038-1043. |
| Liu XT, Wu MC, Yin X, et al. Effect of disulfide bridge on thermostability improvement of the xylanase AoXyn11A[J]. Journal of Food Science and Biotechnology, 2014, 33(10): 1038-1043. | |
| [22] | 李培, 刘欣, 向彬彬, 等. 基于功能纳米材料的蛋白激酶活性分析新方法[J]. 中国科学:化学, 2015, 45(11): 1178-1193. |
|
Li P, Liu X, Xiang BB, et al. A new method for protein kinase activity analysis based on functional nanomaterials[J]. Scientia Sinica:Chimica, 2015, 45(11): 1178-1193.
doi: 10.1360/N032015-00086 URL |
| [1] | WANG Shuai, FENG Yu-mei, BAI Miao, DU Wei-jun, YUE Ai-qin. Functional Analysis of Soybean Gene GmHMGR Responding to Exogenous Hormones and Abiotic Stresses [J]. Biotechnology Bulletin, 2023, 39(7): 131-142. |
| [2] | ZHANG Lu-yang, HAN Wen-long, XU Xiao-wen, YAO Jian, LI Fang-fang, TIAN Xiao-yuan, ZHANG Zhi-qiang. Identification and Expression Analysis of the Tobacco TCP Gene Family [J]. Biotechnology Bulletin, 2023, 39(6): 248-258. |
| [3] | LI Jing-rui, WANG Yu-bo, XIE Zi-wei, LI Chang, WU Xiao-lei, GONG Bin-bin, GAO Hong-bo. Identification and Expression Analysis of PIN Gene Family in Melon Under High Temperature Stress [J]. Biotechnology Bulletin, 2023, 39(5): 192-204. |
| [4] | ZHAO Sai-sai, ZHANG Xiao-dan, JIA Xiao-yan, TAO Da-wei, LIU Ke-yu, NING Xi-bin. Investigation on the Complex Mutagenesis Selection of High-yield Nitrate Reductase Strain Staphylococcus simulans ZSJ6 and Its Enzymatic Properties [J]. Biotechnology Bulletin, 2023, 39(4): 103-113. |
| [5] | MA Yu-qian, SUN Dong-hui, YUE Hao-feng, XIN Jia-yu, LIU Ning, CAO Zhi-yan. Identification, Heterologous Expression and Functional Analysis of a GH61 Family Glycoside Hydrolase from Setosphaeria turcica with the Assisting Function in Degrading Cellulose [J]. Biotechnology Bulletin, 2023, 39(4): 124-135. |
| [6] | GUO San-bao, SONG Mei-ling, LI Ling-xin, YAO Zi-zhao, GUI Ming-ming, HUANG Sheng-he. Cloning and Analysis of Chalcone Synthase Gene and Its Promoter from Euphorbia maculata [J]. Biotechnology Bulletin, 2023, 39(4): 148-156. |
| [7] | WANG Yi-qing, WANG Tao, WEI Chao-ling, DAI Hao-min, CAO Shi-xian, SUN Wei-jiang, ZENG Wen. Identification and Interaction Analysis of SMAS Gene Family in Tea Plant(Camellia sinensis) [J]. Biotechnology Bulletin, 2023, 39(4): 246-258. |
| [8] | CHEN Nan-nan, WANG Chun-lai, JIANG Zhen-zhong, JIAO Peng, GUAN Shu-yan, MA Yi-yong. Genetic Transformation and Chilling Resistance Analysis of Maize ZmDHN15 Gene in Tobacco [J]. Biotechnology Bulletin, 2023, 39(4): 259-267. |
| [9] | YANG Lan, ZHANG Chen-xi, FAN Xue-wei, WANG Yang-guang, WANG Chun-xiu, LI Wen-ting. Gene Cloning, Expression Pattern, and Promoter Activity Analysis of Chicken BMP15 [J]. Biotechnology Bulletin, 2023, 39(4): 304-312. |
| [10] | CHEN Qiang, ZHOU Ming-kang, SONG Jia-min, ZHANG Chong, WU Long-kun. Identification and Analysis of LBD Gene Family and Expression Analysis of Fruit Development in Cucumis melo [J]. Biotechnology Bulletin, 2023, 39(3): 176-183. |
| [11] | PING Huai-lei, GUO Xue, YU Xiao, SONG Jing, DU Chun, WANG Juan, ZHANG Huai-bi. Cloning and Expression of PdANS in Paeonia delavayi and Correlation with Anthocyanin Content [J]. Biotechnology Bulletin, 2023, 39(3): 206-217. |
| [12] | MIAO Shu-nan, GAO Yu, LI Xin-ru, CAI Gui-ping, ZHANG Fei, XUE Jin-ai, JI Chun-li, LI Run-zhi. Functional Analysis of Soybean GmPDAT1 Genes in the Oil Biosynthesis and Response to Abiotic Stresses [J]. Biotechnology Bulletin, 2023, 39(2): 96-106. |
| [13] | XING Yuan, SONG Jian, LI Jun-yi, ZHENG Ting-ting, LIU Si-chen, QIAO Zhi-jun. Identification of AP Gene Family and Its Response Analysis to Abiotic Stress in Setaria italica [J]. Biotechnology Bulletin, 2023, 39(11): 238-251. |
| [14] | CHEN Chu-yi, YANG Xiao-mei, CHEN Sheng-yan, CHEN Bin, YUE Li-ran. Expression Analysis of the ZF-HD Gene Family in Chrysanthemum nankingense Under Drought and ABA Treatment [J]. Biotechnology Bulletin, 2023, 39(11): 270-282. |
| [15] | YANG Min, LONG Yu-qing, ZENG Juan, ZENG Mei, ZHOU Xin-ru, WANG Ling, FU Xue-sen, ZHOU Ri-bao, LIU Xiang-dan. Cloning and Function Analysis of Gene UGTPg17 and UGTPg36 in Lonicera macranthoides [J]. Biotechnology Bulletin, 2023, 39(10): 256-267. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||