








Biotechnology Bulletin ›› 2021, Vol. 37 ›› Issue (11): 267-275.doi: 10.13560/j.cnki.biotech.bull.1985.2020-1506
Previous Articles Next Articles
HE Jin-hua1( ), MA Xiang1, TANG Yan-qiong1, WANG Dan2, LI Hong1(
), MA Xiang1, TANG Yan-qiong1, WANG Dan2, LI Hong1( ), LIU Zhu1
), LIU Zhu1
Received:2020-12-13
Online:2021-11-26
Published:2021-12-03
Contact:
LI Hong
E-mail:hejinhua0625@163.com;lihongbio@163.com
HE Jin-hua, MA Xiang, TANG Yan-qiong, WANG Dan, LI Hong, LIU Zhu. Identification and Function Study of a New Type of sRNA N155 from Aeromonas veronii[J]. Biotechnology Bulletin, 2021, 37(11): 267-275.
| 菌株/质粒 Strain/plasmid | 相关属性 Related attributes | 来源 Sources |
|---|---|---|
| A.veronii C4 | 野生型菌株,具有氨苄青霉素抗性 Wild type strain,having ampicillin resistance | 本实验室 Our lab stock |
| A.veronii Δhfq | hfq基因敲除型菌株,具有氨苄青霉素抗性 Gene hfq-deleted strain,having ampicillin resistance | 本实验室 Our lab stock |
| A.veronii Δhfq-pBBR-hfq | hfq基因回补菌株,具有氨苄青霉素和卡纳霉素双抗性 Gene hfq-complemented strain,having ampicillin and kanamycin resistances | 本研究 This study |
| A.veronii ΔsRNA N155 | sRNA N155敲除型菌株,具有氨苄青霉素抗性 Gene sRNA N155-deleted strain,having ampicillin resistance | 本研究 This study |
| Escherichia coli WM3064 | 二氨基庚二酸(DAP)营养缺陷型菌株 Diaminopimelic acid(DAP)auxotrophic strain | 本实验室 Our lab stock |
| pBBR-MCS-2 | 穿梭质粒,具有卡纳霉素抗性 Broad-host-range cloning vector,having kanamycin resistance | 本实验室 Our lab stock |
| pBBR-hfq | pBBR-MCS-2的衍生质粒,包含hfq的完整ORF区域 Derivatives from pBBR-MCS-2,containing full ORF region of hfq | 本研究 This study |
| pRE112 | 基因敲除质粒,自杀性质粒,表达了用于蔗糖选择的sacB基因,具有氯霉素抗性 Plasmid for gene knock out,suicide plasmid,expressing the sacB gene for sucrose selection,having chloramphenicol resistance | 本实验室 Our lab stock |
| pRE112-ΔsRNA N155 | sRNA N155敲除重组质粒 sRNA N155 knockout recombinant plasmid | 本研究 This study |
Table 1 Strains,plasmids used in this study
| 菌株/质粒 Strain/plasmid | 相关属性 Related attributes | 来源 Sources |
|---|---|---|
| A.veronii C4 | 野生型菌株,具有氨苄青霉素抗性 Wild type strain,having ampicillin resistance | 本实验室 Our lab stock |
| A.veronii Δhfq | hfq基因敲除型菌株,具有氨苄青霉素抗性 Gene hfq-deleted strain,having ampicillin resistance | 本实验室 Our lab stock |
| A.veronii Δhfq-pBBR-hfq | hfq基因回补菌株,具有氨苄青霉素和卡纳霉素双抗性 Gene hfq-complemented strain,having ampicillin and kanamycin resistances | 本研究 This study |
| A.veronii ΔsRNA N155 | sRNA N155敲除型菌株,具有氨苄青霉素抗性 Gene sRNA N155-deleted strain,having ampicillin resistance | 本研究 This study |
| Escherichia coli WM3064 | 二氨基庚二酸(DAP)营养缺陷型菌株 Diaminopimelic acid(DAP)auxotrophic strain | 本实验室 Our lab stock |
| pBBR-MCS-2 | 穿梭质粒,具有卡纳霉素抗性 Broad-host-range cloning vector,having kanamycin resistance | 本实验室 Our lab stock |
| pBBR-hfq | pBBR-MCS-2的衍生质粒,包含hfq的完整ORF区域 Derivatives from pBBR-MCS-2,containing full ORF region of hfq | 本研究 This study |
| pRE112 | 基因敲除质粒,自杀性质粒,表达了用于蔗糖选择的sacB基因,具有氯霉素抗性 Plasmid for gene knock out,suicide plasmid,expressing the sacB gene for sucrose selection,having chloramphenicol resistance | 本实验室 Our lab stock |
| pRE112-ΔsRNA N155 | sRNA N155敲除重组质粒 sRNA N155 knockout recombinant plasmid | 本研究 This study |
| 引物 Primer | 引物序列Primer sequence(5'-3') | 注释 Note |
|---|---|---|
| F0 | GAGCTGGTCTTTATGCGCC | 验证sRNA N155敲除 Validation of sRNA N155 knockout |
| R0 | GGAAGAAAAAGCTGACGAAG | |
| F1 | CGAGCTCCCTGTCTATTGGCACTGC | 扩增sRNA N155上游同源序列 Amplification of sRNA N155 upstream homologous sequence |
| R1 | CCGGAATTCGAACAGGTTGCAGAAGTC | |
| F2 | CGGAATTCTTTACTGCGCAGTGTTGAATC | 扩增sRNA N155下游同源序列 Amplification of sRNA N155 downstream homologous sequence |
| R2 | CGGGGTACCAATCGCAGCTGTTACAAAG | |
| pRE112 F | ACATAGCCCCACTGTTCGT | 验证pRE112载体 Verification of pRE112 vector |
| pRE112 R | TTTTCGTCTCAGCCAATCC | |
| GyrB F | TGGTTGTGGTATCGGTCGTG | 内参基因引物 Primers for reference gene |
| GyrB R | CTGTTCCTGCTTGCCTTT | |
| RT-F | TTATCCGGTAAGGAGACG | sRNA N155 RT-qPCR引物 sRNA N155 RT-qPCR primers |
| RT-R | CACGCGGTAAAAGAACAC |
Table 2 Primer sequence used in this study
| 引物 Primer | 引物序列Primer sequence(5'-3') | 注释 Note |
|---|---|---|
| F0 | GAGCTGGTCTTTATGCGCC | 验证sRNA N155敲除 Validation of sRNA N155 knockout |
| R0 | GGAAGAAAAAGCTGACGAAG | |
| F1 | CGAGCTCCCTGTCTATTGGCACTGC | 扩增sRNA N155上游同源序列 Amplification of sRNA N155 upstream homologous sequence |
| R1 | CCGGAATTCGAACAGGTTGCAGAAGTC | |
| F2 | CGGAATTCTTTACTGCGCAGTGTTGAATC | 扩增sRNA N155下游同源序列 Amplification of sRNA N155 downstream homologous sequence |
| R2 | CGGGGTACCAATCGCAGCTGTTACAAAG | |
| pRE112 F | ACATAGCCCCACTGTTCGT | 验证pRE112载体 Verification of pRE112 vector |
| pRE112 R | TTTTCGTCTCAGCCAATCC | |
| GyrB F | TGGTTGTGGTATCGGTCGTG | 内参基因引物 Primers for reference gene |
| GyrB R | CTGTTCCTGCTTGCCTTT | |
| RT-F | TTATCCGGTAAGGAGACG | sRNA N155 RT-qPCR引物 sRNA N155 RT-qPCR primers |
| RT-R | CACGCGGTAAAAGAACAC |

Fig. 6 cDNA agarose gel electrophoresis M:DL5000 DNA marker. 1-2 are the bands of internal reference gene GyrB of Escherichia coli of WT+pACYC. 3-4 are the bands of internal reference gene GyrB of WT+ pACYCDuet-1∷psRNAN155-sRNA N155. 5-6 are RT products of sRNA N155 of WT+pACYC. 7-8 are RT products of sRNA N155 of WT+pACYCDuet-1∷psRNAN155-sRNA N155,about 110 bp
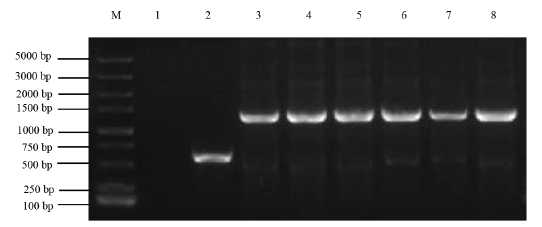
Fig. 8 PCR verification of sRNA N155 gene knockout vector pRE112-ΔsRNA N155 M:DL5000 DNA marker. 1 is a negative control. 2 is a positive control. 3-8 are plasmid PCR products of pRE112-ΔsRNA N155 gene knockout,the target band is about 1 200 bp

Fig. 9 22% sucrose plate screening M:DL5000 DNA marker. 1:A. veronii C4 genome positive control. 22:pRE112 plasmid positive control. 2-13:Knockout strain verification,where 2-3,10-11 are the same as expected,all are lower than the wild type. 14-21 are pRE112-specific primer verification,indicating that the pRE112 plasmid has been successfully lost

Fig. 10 PCR verification of A.veronii sRNA N155 knockout strain M:DL5000 DNA marker. 1:A.veronii C4 genome positive control. 2-5:sRNA N155 knockout strain PCR verification

Fig. 12 Effect of knocking out the sRNA N155 gene on bacterial motility 1:A.veronii C4 wild-type strain. 2:Δhfq knockout strain. 3:ΔsRNA N155 knockout strain. 4:Δhfq-pBBR-hfq complement strain
| [1] |
Pablos M, Huys G, Huys G, et al. Identification and epidemiological relationships of Aeromonas isolates from patients with diarrhea, drinking water and foods[J]. Int J Food Microbiol, 2011, 147(3): 203-210.
doi: 10.1016/j.ijfoodmicro.2011.04.006 pmid: 21550680 |
| [2] | 敬小兵, 刘天强, 黄冠军, 等. 一种检测维氏气单胞菌的试剂盒和方法:中国, 105506153A[P]. 2018-12-28. |
| Jing XB, Liu TQ, Huang GJ, et al. A kit and method for the detection of Aeromonas veronii: China, 105506153A[P]. 2018-12-28. | |
| [3] | 葛铮. 不同淡水鱼类维氏气单胞菌的分离鉴定及耐药性分析[D]. 长春:吉林农业大学, 2015. |
| Ge Z. Isolated identification and drug resistance analysis of Aeromonas veronii from different freshwater fish species[D]. Changchun:Jilin Agricultural University, 2015. | |
| [4] |
Fang HM, Ling KC, Ge R, et al. Enhancement of protective immunity in blue gourami, Trichogaster trichopterus(Pallas), against Aer-omonas hydrophila and Vibrio anguillarum by A. hydrophila major adhesin[J]. Journal of Fish Diseases, 2000, 23(2): 137-145.
doi: 10.1046/j.1365-2761.2000.00229.x URL |
| [5] | 康元环, 张冬星, 杨滨僮, 等. 维氏气单胞菌最新研究进展[J]. 中国人兽共患病学报, 2018, 34(5): 452-459, 465. |
| Kang YH, Zhang DX, Yang BT, et al. Latest research progress on Aeromonas veronii[J]. Chin J Zoonoses, 2018, 34(5): 452-459, 465. | |
| [6] |
Jeamton W, Mungpakdee S, Sirijuntarut M, et al. A combined stress response analysis of Spirulina platensis in terms of global differentially expressed proteins, and mRNA levels and stability of fatty acid biosynjournal genes[J]. FEMS Microbiol Lett, 2008, 281(2): 121-131.
doi: 10.1111/fml.2008.281.issue-2 URL |
| [7] | 赵小凯, 竹俊兰, 严浩, 等. 细菌sRNA功能、预测及鉴定方法的研究进展[J]. 温州医学院学报, 2012, 42(5): 503-507. |
| Zhao XK, Zhu JL, Yan H, et al. Research progress in bacterial sRNA function, prediction and identification methods[J]. J Wenzhou Med Coll, 2012, 42(5): 503-507. | |
| [8] |
Morita T, Nishino R, Aiba H. Role of the Terminator hairpin in the biogenesis of functional Hfq-binding sRNAs[J]. RNA, 2017, 23(9): 1419-1431.
doi: 10.1261/rna.060756.117 URL |
| [9] |
Zhang S, Liu S, Wu N, et al. Small non-coding RNA RyhB mediates persistence to multiple antibiotics and stresses in uropathogenic Escherichia coli by reducing cellular metabolism[J]. Front Microbiol, 2018, 9: 136.
doi: 10.3389/fmicb.2018.00136 URL |
| [10] |
Jørgensen MG, Thomason MK, Havelund J, et al. Dual function of the McaS small RNA in controlling biofilm formation[J]. Genes Dev, 2013, 27(10): 1132-1145.
doi: 10.1101/gad.214734.113 URL |
| [11] |
Parker A, Cureoglu S, De Lay N, et al. Alternative pathways for Escherichia coli biofilm formation revealed by sRNA overproduction[J]. Mol Microbiol, 2017, 105(2): 309-325.
doi: 10.1111/mmi.2017.105.issue-2 URL |
| [12] |
Li MY, Zhang J, Lu P, et al. Evaluation of biological characteristics of bacteria contributing to biofilm formation[J]. Pedosphere, 2009, 19(5): 554-561.
doi: 10.1016/S1002-0160(09)60149-1 URL |
| [13] |
Urbanowski ML, Stauffer LT, Stauffer GV. The gcvB gene encodes a small untranslated RNA involved in expression of the dipeptide and oligopeptide transport systems in Escherichia coli[J]. Mol Microbiol, 2000, 37(4): 856-868.
pmid: 10972807 |
| [14] |
Heidrich N, Chinali A, Gerth U, et al. The small untranslated RNA SR1 from the Bacillus subtilis genome is involved in the regulation of arginine catabolism[J]. Mol Microbiol, 2006, 62(2): 520-536.
pmid: 17020585 |
| [15] |
Gerrick ER, Barbier T, Chase MR, et al. Small RNA profiling in Mycobacterium tuberculosis identifies MrsI as necessary for an anticipatory iron sparing response[J]. PNAS, 2018, 115(25): 6464-6469.
doi: 10.1073/pnas.1718003115 pmid: 29871950 |
| [16] |
Massé E, Salvail H, Desnoyers G, et al. Small RNAs controlling iron metabolism[J]. Curr Opin Microbiol, 2007, 10(2): 140-145.
pmid: 17383226 |
| [17] |
Johnson JR, Clabots C, Rosen H. Effect of inactivation of the global oxidative stress regulator oxyR on the colonization ability of Escherichia coli O1:K1:H7 in a mouse model of ascending urinary tract infection[J]. Infect Immun, 2006, 74(1): 461-468.
pmid: 16369002 |
| [18] | Wagner EGH, Romby P. Small RNAs in bacteria and archaea:who they are, what they do, and how they do it[J]. Adv Genet, 2015, 90: 133-208. |
| [19] |
Yu WJ, Li DY, Li H, et al. Absence of tmRNA increases the persistence to cefotaxime and the intercellular accumulation of metabolite GlcNAc in Aeromonas veronii[J]. Front Cell Infect Microbiol, 2020, 10: 44.
doi: 10.3389/fcimb.2020.00044 URL |
| [20] |
Kong L, Zhang Y, Ye ZQ, et al. CPC:assess the protein-coding potential of transcripts using sequence features and support vector machine[J]. Nucleic Acids Res, 2007, 35(web server issue): W345-W349.
doi: 10.1093/nar/gkm391 URL |
| [21] |
Kumar L, E Futschik M. Mfuzz:a software package for soft clustering of microarray data[J]. Bioinformation, 2007, 2(1): 5-7.
doi: 10.6026/bioinformation URL |
| [22] |
Zhang QY, Wang Q, Ouyang HL, et al. Pyrosequencing reveals significant changes in microbial communities along the ecological succession of biological soil crusts in the tengger desert of China[J]. Pedosphere, 2018, 28(2): 350-362.
doi: 10.1016/S1002-0160(17)60477-6 URL |
| [1] | LIN Hong-yan, GUO Xiao-rui, LIU Di, LI Hui, LU Hai. Molecular Mechanism of Transcriptional Factor AtbHLH68 in Regulating Cell Wall Development by Transcriptome Analysis [J]. Biotechnology Bulletin, 2023, 39(9): 105-116. |
| [2] | MIAO Yong-mei, MIAO Cui-ping, YU Qing-cai. Properties of Bacillus subtilis Strain BBs-27 Fermentation Broth and the Inhibition of Lipopeptides Against Fusarium culmorum [J]. Biotechnology Bulletin, 2023, 39(9): 255-267. |
| [3] | FU Yu, JIA Rui-rui, HE He, WANG Liang-gui, YANG Xiu-lian. Growth Differences Among Grafted Seedlings with Two Rootstocks of Catalpa bungei and Comparative Analysis of Transcriptome [J]. Biotechnology Bulletin, 2023, 39(8): 251-261. |
| [4] | KONG De-zhen, DUAN Zhen-yu, WANG Gang, ZHANG Xin, XI Lin-qiao. Physiological Characteristics and Transcriptome Analysis of Sorghum bicolor × S. Sudanense Seedlings Under Salt-alkali Stress [J]. Biotechnology Bulletin, 2023, 39(6): 199-207. |
| [5] | LIU Hui, LU Yang, YE Xi-miao, ZHOU Shuai, LI Jun, TANG Jian-bo, CHEN En-fa. Comparative Transcriptome Analysis of Cadmium Stress Response Induced by Exogenous Sulfur in Tartary Buckwheat [J]. Biotechnology Bulletin, 2023, 39(5): 177-191. |
| [6] | XIE Yang, XING Yu-meng, ZHOU Guo-yan, LIU Mei-yan, YIN Shan-shan, YAN Li-ying. Transcriptome Analysis of Diploid and Autotetraploid in Cucumber Fruit [J]. Biotechnology Bulletin, 2023, 39(3): 152-162. |
| [7] | HU Li-li, LIN Bo-rong, WANG Hong-hong, CHEN Jian-song, LIAO Jin-ling, ZHUO Kan. Transcriptome and Candidate Effectors Analysis of Pratylenchus brachyurus [J]. Biotechnology Bulletin, 2023, 39(3): 254-266. |
| [8] | SUN Yan-qiu, XIE Cai-yun, TANG Yue-qin. Construction and Mechanism Analysis of High-temperature Resistant Saccharomyces cerevisiae [J]. Biotechnology Bulletin, 2023, 39(11): 226-237. |
| [9] | XU Jun, YE Yu-qing, NIU Ya-jing, HUANG He, ZHANG Meng-meng. Transcriptome Analysis of Rhizome Development in Chrysanthemum× × morifolium [J]. Biotechnology Bulletin, 2023, 39(10): 231-245. |
| [10] | LUO Hao-tian, WANG Long, WANG Yu-qian, WANG Yue, LI Jia-zhen, YANG Meng-ke, ZHANG Jie, DENG Xin, WANG Hong-yan. Genome-wide Identification and Expression Analysis of the RNAi-related Gene Families in Setaria viridis [J]. Biotechnology Bulletin, 2023, 39(1): 175-186. |
| [11] | XIN Jian-pan, LI Yan, ZHAO Chu, TIAN Ru-nan. Transcriptome Sequencing in the Leaves of Pontederia cordata with Cadmium Exposure and Gene Mining in Phenypropanoid Pathways [J]. Biotechnology Bulletin, 2022, 38(6): 198-210. |
| [12] | XU Jin, LI Tao, LI Chu-lin, ZHU Shun-ni, WANG Zhong-ming, XIANG Wen-zhou. Effects of Temperature on the Growth,Total Lipid and Eicosapentaenoic Acid Synthesis of Eustigmatos sp. [J]. Biotechnology Bulletin, 2022, 38(6): 261-271. |
| [13] | XIONG He-li, SHA Qian, LIU Shao-na, XIANG De-cai, ZHANG Bin, ZHAO Zhi-yong. Application of Single-cell Transcriptome Sequencing in Animals [J]. Biotechnology Bulletin, 2022, 38(3): 226-233. |
| [14] | ZHANG Bin, YANG Xin-xia. Identification of Key Transcription Factors in Response to Salt Stress in Rice [J]. Biotechnology Bulletin, 2022, 38(3): 9-15. |
| [15] | GUAN Yi, LI Xin, WANG Ding-yi, DU Xi, ZHANG Long-bin, YE Xiu-yun. Functional Study of BbRho5 on the Growth Rate of Beauveria bassiana [J]. Biotechnology Bulletin, 2022, 38(2): 132-140. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||