








Biotechnology Bulletin ›› 2022, Vol. 38 ›› Issue (2): 21-33.doi: 10.13560/j.cnki.biotech.bull.1985.2020-1327
Previous Articles Next Articles
FENG Jian-ying( ), LI Li-qin, LU Li-ming(
), LI Li-qin, LU Li-ming( )
)
Received:2020-10-28
Online:2022-02-26
Published:2022-03-09
Contact:
LU Li-ming
E-mail:1500098916@qq.com;louis_luliming@126.com
FENG Jian-ying, LI Li-qin, LU Li-ming. Genome-wide Identification and Expression Analysis of the bHLH Transcription Factor Family in Solanum tuberosum[J]. Biotechnology Bulletin, 2022, 38(2): 21-33.
| 基因 Gene | 上游引物 Forward primer(5'-3') | 下游引物 Reverse primer(5'-3') |
|---|---|---|
| StbHLH2 | ATGTTCACCGCCTTATTG | CTGATGAGATTAGAGCCACC |
| StbHLH14 | CTAAAGGTGGAAGTGGGGTA | TTGGGGGTTGTCAATAAAAT |
| StbHLH15 | ACAAGTTGCCCACCTACC | TGGAAGTTCTCACTGCCTCT |
| StbHLH34 | AAAGGAACCGCCGCAAAC | GCGTCTTGGCTTCTAAGGATT |
| StbHLH39 | ATCCTCTTGCTCGCCTGTT | GGAATAGTAGATTTTTGGGGCA |
| StbHLH53 | ACCTCCTGTTCCGATGAT | ACCAAAGTTCCCTCCACT |
| StbHLH57 | CATTGGATAAAGTCACCGATAC | CGAATCTACAGACGACCTCA |
| StbHLH59 | GGCATTTTTCACGGTTGCCT | CCGTCGTCACTTCCCTATCG |
| StbHLH66 | AGCACTTGTTCCTGGTTGTGA | AATGGTCCTTCCAAGCCACTC |
| StbHLH80 | CCAGATTCACCCCGTCGTTT | CCTTGTGCTCACGAAACACC |
| StbHLH90 | CCAGTTCCACTACACCGTCC | GCCGGAGGATTTGACGGAAT |
| StbHLH108 | GTCAAATCGAAGAACACGCGG | CTGCGGCTTTCAGGGATAAGT |
Table 1 Primer sequences
| 基因 Gene | 上游引物 Forward primer(5'-3') | 下游引物 Reverse primer(5'-3') |
|---|---|---|
| StbHLH2 | ATGTTCACCGCCTTATTG | CTGATGAGATTAGAGCCACC |
| StbHLH14 | CTAAAGGTGGAAGTGGGGTA | TTGGGGGTTGTCAATAAAAT |
| StbHLH15 | ACAAGTTGCCCACCTACC | TGGAAGTTCTCACTGCCTCT |
| StbHLH34 | AAAGGAACCGCCGCAAAC | GCGTCTTGGCTTCTAAGGATT |
| StbHLH39 | ATCCTCTTGCTCGCCTGTT | GGAATAGTAGATTTTTGGGGCA |
| StbHLH53 | ACCTCCTGTTCCGATGAT | ACCAAAGTTCCCTCCACT |
| StbHLH57 | CATTGGATAAAGTCACCGATAC | CGAATCTACAGACGACCTCA |
| StbHLH59 | GGCATTTTTCACGGTTGCCT | CCGTCGTCACTTCCCTATCG |
| StbHLH66 | AGCACTTGTTCCTGGTTGTGA | AATGGTCCTTCCAAGCCACTC |
| StbHLH80 | CCAGATTCACCCCGTCGTTT | CCTTGTGCTCACGAAACACC |
| StbHLH90 | CCAGTTCCACTACACCGTCC | GCCGGAGGATTTGACGGAAT |
| StbHLH108 | GTCAAATCGAAGAACACGCGG | CTGCGGCTTTCAGGGATAAGT |
| 基因 Gene | 基因ID Gene ID | 氨基酸数Number of amino acid | 分子量 Molecular weight/Da | 等电点 pI | 基因 Gene | 基因ID Gene ID | 氨基酸数Number of amino acid | 分子量 Molecular weight/Da | 等电点 pI | |
|---|---|---|---|---|---|---|---|---|---|---|
| StbHLH1 | PGSC0003DMT400095438 | 295 | 33 008.42 | 8.21 | StbHLH39 | PGSC0003DMT400036951 | 551 | 59 473.80 | 5.78 | |
| StbHLH2 | PGSC0003DMT400065029 | 397 | 43 515.43 | 6.16 | StbHLH40 | PGSC0003DMT400014656 | 276 | 31 123.90 | 5.76 | |
| StbHLH3 | PGSC0003DMT400004497 | 471 | 53 156.01 | 5.18 | StbHLH41 | PGSC0003DMT400006783 | 311 | 34 469.88 | 6.45 | |
| StbHLH4 | PGSC0003DMT400086863 | 393 | 44 510.14 | 6.01 | StbHLH42 | PGSC0003DMT400015377 | 203 | 22 928.51 | 9.69 | |
| StbHLH5 | PGSC0003DMT400066807 | 464 | 50 747.96 | 7.62 | StbHLH43 | PGSC0003DMT400060615 | 224 | 25 030.63 | 7.56 | |
| StbHLH6 | PGSC0003DMT400000652 | 598 | 66 369.87 | 6.86 | StbHLH44 | PGSC0003DMT400027293 | 235 | 27 571.74 | 8.82 | |
| StbHLH7 | PGSC0003DMT400047220 | 452 | 49 967.36 | 5.61 | StbHLH45 | PGSC0003DMT400005446 | 345 | 38 526.03 | 5.87 | |
| StbHLH8 | PGSC0003DMT400073574 | 337 | 38 301.38 | 5.14 | StbHLH46 | PGSC0003DMT400033589 | 321 | 35 634.69 | 4.55 | |
| StbHLH9 | PGSC0003DMT400013074 | 353 | 39 806.96 | 6.33 | StbHLH47 | PGSC0003DMT400024185 | 204 | 22 750.35 | 9.88 | |
| StbHLH10 | PGSC0003DMT400087143 | 333 | 36 913.79 | 5.28 | StbHLH48 | PGSC0003DMT400016360 | 338 | 37 784.66 | 4.66 | |
| StbHLH11 | PGSC0003DMT400066257 | 305 | 34 150.70 | 7.11 | StbHLH49 | PGSC0003DMT400012591 | 382 | 41 627.38 | 6.33 | |
| StbHLH12 | PGSC0003DMT400066259 | 307 | 34 354.03 | 6.67 | StbHLH50 | PGSC0003DMT400020534 | 314 | 34 098.61 | 5.32 | |
| StbHLH13 | PGSC0003DMT400066261 | 304 | 33 738.26 | 7.14 | StbHLH51 | PGSC0003DMT400009603 | 406 | 44 073.59 | 7.66 | |
| StbHLH14 | PGSC0003DMT400066264 | 307 | 34 179.83 | 7.12 | StbHLH52 | PGSC0003DMT400050572 | 321 | 35 928.50 | 5.11 | |
| StbHLH15 | PGSC0003DMT400001888 | 442 | 49 692.41 | 5.65 | StbHLH53 | PGSC0003DMT400050585 | 520 | 56 513.05 | 5.21 | |
| StbHLH16 | PGSC0003DMT400040021 | 303 | 34 207.55 | 6.54 | StbHLH54 | PGSC0003DMT400002155 | 312 | 34 997.42 | 6.00 | |
| StbHLH17 | PGSC0003DMT400026937 | 326 | 36 823.76 | 5.23 | StbHLH55 | PGSC0003DMT400033757 | 210 | 23 748.34 | 8.89 | |
| StbHLH18 | PGSC0003DMT400021383 | 244 | 27 458.61 | 5.56 | StbHLH56 | PGSC0003DMT400044959 | 231 | 26 357.79 | 5.64 | |
| StbHLH19 | PGSC0003DMT400026449 | 235 | 26 275.12 | 8.64 | StbHLH57 | PGSC0003DMT400018046 | 578 | 64 285.91 | 7.61 | |
| StbHLH20 | PGSC0003DMT400036021 | 442 | 49 217.42 | 5.90 | StbHLH58 | PGSC0003DMT400040761 | 390 | 44 069.48 | 5.27 | |
| StbHLH21 | PGSC0003DMT400055910 | 267 | 29 914.13 | 7.80 | StbHLH59 | PGSC0003DMT400038134 | 515 | 56 408.13 | 5.13 | |
| StbHLH22 | PGSC0003DMT400078776 | 317 | 35 038.85 | 6.12 | StbHLH60 | PGSC0003DMT400048941 | 312 | 35 241.59 | 4.72 | |
| StbHLH23 | PGSC0003DMT400078675 | 335 | 38 417.26 | 5.44 | StbHLH61 | PGSC0003DMT400040448 | 225 | 25 610.16 | 7.62 | |
| StbHLH24 | PGSC0003DMT400078696 | 347 | 39 529.70 | 6.39 | StbHLH62 | PGSC0003DMT400071861 | 62 | 7 527.78 | 10.4 | |
| StbHLH25 | PGSC0003DMT400078698 | 209 | 23 532.27 | 6.17 | StbHLH63 | PGSC0003DMT400080418 | 260 | 28 511.99 | 7.81 | |
| StbHLH26 | PGSC0003DMT400078699 | 323 | 35 931.15 | 5.47 | StbHLH64 | PGSC0003DMT400012487 | 390 | 42 992.79 | 5.76 | |
| StbHLH27 | PGSC0003DMT400010253 | 376 | 41 285.79 | 5.04 | StbHLH65 | PGSC0003DMT400074075 | 530 | 57 936.63 | 5.31 | |
| StbHLH28 | PGSC0003DMT400034838 | 296 | 32 476.68 | 5.27 | StbHLH66 | PGSC0003DMT400083164 | 416 | 46 807.69 | 8.71 | |
| StbHLH29 | PGSC0003DMT400012997 | 335 | 38 164.49 | 5.23 | StbHLH66 | PGSC0003DMT400069569 | 284 | 31 977.18 | 6.76 | |
| StbHLH30 | PGSC0003DMT400058169 | 268 | 30 772.88 | 7.63 | StbHLH68 | PGSC0003DMT400051980 | 467 | 51 763.96 | 7.19 | |
| StbHLH31 | PGSC0003DMT400048857 | 148 | 16 910.86 | 9.75 | StbHLH69 | PGSC0003DMT400051674 | 477 | 52 903.87 | 6.59 | |
| StbHLH32 | PGSC0003DMT400071293 | 224 | 25 544.12 | 8.60 | StbHLH70 | PGSC0003DMT400028917 | 444 | 46 271.20 | 6.09 | |
| StbHLH33 | PGSC0003DMT400019550 | 214 | 24 405.08 | 8.94 | StbHLH71 | PGSC0003DMT400010463 | 252 | 28 629.96 | 4.97 | |
| StbHLH34 | PGSC0003DMT400023497 | 355 | 38 301.12 | 5.94 | StbHLH72 | PGSC0003DMT400056685 | 374 | 42 308.55 | 8.80 | |
| StbHLH35 | PGSC0003DMT400051137 | 338 | 37 637.08 | 6.08 | StbHLH73 | PGSC0003DMT400041156 | 517 | 57 437.21 | 7.09 | |
| StbHLH36 | PGSC0003DMT400062844 | 291 | 30 828.06 | 5.65 | StbHLH74 | PGSC0003DMT400067287 | 351 | 39 693.02 | 8.14 | |
| StbHLH37 | PGSC0003DMT400063026 | 536 | 58 417.99 | 7.35 | StbHLH75 | PGSC0003DMT400052622 | 255 | 27 880.55 | 6.92 | |
| StbHLH38 | PGSC0003DMT400063117 | 437 | 48 174.88 | 6.79 | StbHLH76 | PGSC0003DMT400018274 | 224 | 25 831.73 | 6.32 | |
| StbHLH77 | PGSC0003DMT400049504 | 485 | 52 745.68 | 6.07 | StbHLH93 | PGSC0003DMT400016793 | 348 | 36 847.65 | 8.57 | |
| StbHLH78 | PGSC0003DMT400002963 | 598 | 65 258.62 | 5.92 | StbHLH94 | PGSC0003DMT400064399 | 388 | 42 575.81 | 5.83 | |
| StbHLH79 | PGSC0003DMT400015017 | 456 | 51 791.64 | 6.22 | StbHLH95 | PGSC0003DMT400028057 | 389 | 44 514.29 | 5.88 | |
| StbHLH80 | PGSC0003DMT400021165 | 435 | 49 379.65 | 5.89 | StbHLH96 | PGSC0003DMT400008370 | 198 | 22 043.97 | 8.88 | |
| StbHLH81 | PGSC0003DMT400053651 | 319 | 36 510.05 | 4.85 | StbHLH97 | PGSC0003DMT400089728 | 305 | 34 320.40 | 5.48 | |
| StbHLH82 | PGSC0003DMT400095303 | 311 | 35 808.26 | 4.90 | StbHLH98 | PGSC0003DMT400076759 | 180 | 20 751.84 | 9.58 | |
| StbHLH83 | PGSC0003DMT400045210 | 330 | 36 732.34 | 6.10 | StbHLH99 | PGSC0003DMT400021130 | 360 | 39 675.21 | 5.92 | |
| StbHLH84 | PGSC0003DMT400045204 | 694 | 75 939.94 | 5.47 | StbHLH100 | PGSC0003DMT400021062 | 380 | 41 565.47 | 9.10 | |
| StbHLH85 | PGSC0003DMT400032139 | 360 | 40 669.07 | 6.81 | StbHLH101 | PGSC0003DMT400041644 | 237 | 27 276.91 | 5.27 | |
| StbHLH86 | PGSC0003DMT400031899 | 431 | 48 790.06 | 6.71 | StbHLH102 | PGSC0003DMT400073760 | 262 | 30 275.48 | 5.87 | |
| StbHLH87 | PGSC0003DMT400053639 | 492 | 53 992.33 | 7.13 | StbHLH103 | PGSC0003DMT400056716 | 441 | 48 896.06 | 6.65 | |
| StbHLH88 | PGSC0003DMT400048756 | 578 | 62 943.67 | 6.42 | StbHLH104 | PGSC0003DMT400020244 | 382 | 39 561.65 | 6.21 | |
| StbHLH89 | PGSC0003DMT400002512 | 284 | 31 495.30 | 5.46 | StbHLH105 | PGSC0003DMT400013766 | 272 | 31 224.18 | 6.76 | |
| StbHLH90 | PGSC0003DMT400033569 | 590 | 64 760.08 | 5.81 | StbHLH106 | PGSC0003DMT400045820 | 296 | 33 899.60 | 7.02 | |
| StbHLH91 | PGSC0003DMT400076223 | 285 | 32 950.42 | 6.27 | StbHLH107 | PGSC0003DMT400011651 | 413 | 46 698.70 | 5.92 | |
| StbHLH92 | PGSC0003DMT400042791 | 439 | 49 242.61 | 6.06 | StbHLH108 | PGSC0003DMT400061284 | 489 | 54 218.28 | 5.12 |
Table 2 Analysis of basic physical and chemical properties
| 基因 Gene | 基因ID Gene ID | 氨基酸数Number of amino acid | 分子量 Molecular weight/Da | 等电点 pI | 基因 Gene | 基因ID Gene ID | 氨基酸数Number of amino acid | 分子量 Molecular weight/Da | 等电点 pI | |
|---|---|---|---|---|---|---|---|---|---|---|
| StbHLH1 | PGSC0003DMT400095438 | 295 | 33 008.42 | 8.21 | StbHLH39 | PGSC0003DMT400036951 | 551 | 59 473.80 | 5.78 | |
| StbHLH2 | PGSC0003DMT400065029 | 397 | 43 515.43 | 6.16 | StbHLH40 | PGSC0003DMT400014656 | 276 | 31 123.90 | 5.76 | |
| StbHLH3 | PGSC0003DMT400004497 | 471 | 53 156.01 | 5.18 | StbHLH41 | PGSC0003DMT400006783 | 311 | 34 469.88 | 6.45 | |
| StbHLH4 | PGSC0003DMT400086863 | 393 | 44 510.14 | 6.01 | StbHLH42 | PGSC0003DMT400015377 | 203 | 22 928.51 | 9.69 | |
| StbHLH5 | PGSC0003DMT400066807 | 464 | 50 747.96 | 7.62 | StbHLH43 | PGSC0003DMT400060615 | 224 | 25 030.63 | 7.56 | |
| StbHLH6 | PGSC0003DMT400000652 | 598 | 66 369.87 | 6.86 | StbHLH44 | PGSC0003DMT400027293 | 235 | 27 571.74 | 8.82 | |
| StbHLH7 | PGSC0003DMT400047220 | 452 | 49 967.36 | 5.61 | StbHLH45 | PGSC0003DMT400005446 | 345 | 38 526.03 | 5.87 | |
| StbHLH8 | PGSC0003DMT400073574 | 337 | 38 301.38 | 5.14 | StbHLH46 | PGSC0003DMT400033589 | 321 | 35 634.69 | 4.55 | |
| StbHLH9 | PGSC0003DMT400013074 | 353 | 39 806.96 | 6.33 | StbHLH47 | PGSC0003DMT400024185 | 204 | 22 750.35 | 9.88 | |
| StbHLH10 | PGSC0003DMT400087143 | 333 | 36 913.79 | 5.28 | StbHLH48 | PGSC0003DMT400016360 | 338 | 37 784.66 | 4.66 | |
| StbHLH11 | PGSC0003DMT400066257 | 305 | 34 150.70 | 7.11 | StbHLH49 | PGSC0003DMT400012591 | 382 | 41 627.38 | 6.33 | |
| StbHLH12 | PGSC0003DMT400066259 | 307 | 34 354.03 | 6.67 | StbHLH50 | PGSC0003DMT400020534 | 314 | 34 098.61 | 5.32 | |
| StbHLH13 | PGSC0003DMT400066261 | 304 | 33 738.26 | 7.14 | StbHLH51 | PGSC0003DMT400009603 | 406 | 44 073.59 | 7.66 | |
| StbHLH14 | PGSC0003DMT400066264 | 307 | 34 179.83 | 7.12 | StbHLH52 | PGSC0003DMT400050572 | 321 | 35 928.50 | 5.11 | |
| StbHLH15 | PGSC0003DMT400001888 | 442 | 49 692.41 | 5.65 | StbHLH53 | PGSC0003DMT400050585 | 520 | 56 513.05 | 5.21 | |
| StbHLH16 | PGSC0003DMT400040021 | 303 | 34 207.55 | 6.54 | StbHLH54 | PGSC0003DMT400002155 | 312 | 34 997.42 | 6.00 | |
| StbHLH17 | PGSC0003DMT400026937 | 326 | 36 823.76 | 5.23 | StbHLH55 | PGSC0003DMT400033757 | 210 | 23 748.34 | 8.89 | |
| StbHLH18 | PGSC0003DMT400021383 | 244 | 27 458.61 | 5.56 | StbHLH56 | PGSC0003DMT400044959 | 231 | 26 357.79 | 5.64 | |
| StbHLH19 | PGSC0003DMT400026449 | 235 | 26 275.12 | 8.64 | StbHLH57 | PGSC0003DMT400018046 | 578 | 64 285.91 | 7.61 | |
| StbHLH20 | PGSC0003DMT400036021 | 442 | 49 217.42 | 5.90 | StbHLH58 | PGSC0003DMT400040761 | 390 | 44 069.48 | 5.27 | |
| StbHLH21 | PGSC0003DMT400055910 | 267 | 29 914.13 | 7.80 | StbHLH59 | PGSC0003DMT400038134 | 515 | 56 408.13 | 5.13 | |
| StbHLH22 | PGSC0003DMT400078776 | 317 | 35 038.85 | 6.12 | StbHLH60 | PGSC0003DMT400048941 | 312 | 35 241.59 | 4.72 | |
| StbHLH23 | PGSC0003DMT400078675 | 335 | 38 417.26 | 5.44 | StbHLH61 | PGSC0003DMT400040448 | 225 | 25 610.16 | 7.62 | |
| StbHLH24 | PGSC0003DMT400078696 | 347 | 39 529.70 | 6.39 | StbHLH62 | PGSC0003DMT400071861 | 62 | 7 527.78 | 10.4 | |
| StbHLH25 | PGSC0003DMT400078698 | 209 | 23 532.27 | 6.17 | StbHLH63 | PGSC0003DMT400080418 | 260 | 28 511.99 | 7.81 | |
| StbHLH26 | PGSC0003DMT400078699 | 323 | 35 931.15 | 5.47 | StbHLH64 | PGSC0003DMT400012487 | 390 | 42 992.79 | 5.76 | |
| StbHLH27 | PGSC0003DMT400010253 | 376 | 41 285.79 | 5.04 | StbHLH65 | PGSC0003DMT400074075 | 530 | 57 936.63 | 5.31 | |
| StbHLH28 | PGSC0003DMT400034838 | 296 | 32 476.68 | 5.27 | StbHLH66 | PGSC0003DMT400083164 | 416 | 46 807.69 | 8.71 | |
| StbHLH29 | PGSC0003DMT400012997 | 335 | 38 164.49 | 5.23 | StbHLH66 | PGSC0003DMT400069569 | 284 | 31 977.18 | 6.76 | |
| StbHLH30 | PGSC0003DMT400058169 | 268 | 30 772.88 | 7.63 | StbHLH68 | PGSC0003DMT400051980 | 467 | 51 763.96 | 7.19 | |
| StbHLH31 | PGSC0003DMT400048857 | 148 | 16 910.86 | 9.75 | StbHLH69 | PGSC0003DMT400051674 | 477 | 52 903.87 | 6.59 | |
| StbHLH32 | PGSC0003DMT400071293 | 224 | 25 544.12 | 8.60 | StbHLH70 | PGSC0003DMT400028917 | 444 | 46 271.20 | 6.09 | |
| StbHLH33 | PGSC0003DMT400019550 | 214 | 24 405.08 | 8.94 | StbHLH71 | PGSC0003DMT400010463 | 252 | 28 629.96 | 4.97 | |
| StbHLH34 | PGSC0003DMT400023497 | 355 | 38 301.12 | 5.94 | StbHLH72 | PGSC0003DMT400056685 | 374 | 42 308.55 | 8.80 | |
| StbHLH35 | PGSC0003DMT400051137 | 338 | 37 637.08 | 6.08 | StbHLH73 | PGSC0003DMT400041156 | 517 | 57 437.21 | 7.09 | |
| StbHLH36 | PGSC0003DMT400062844 | 291 | 30 828.06 | 5.65 | StbHLH74 | PGSC0003DMT400067287 | 351 | 39 693.02 | 8.14 | |
| StbHLH37 | PGSC0003DMT400063026 | 536 | 58 417.99 | 7.35 | StbHLH75 | PGSC0003DMT400052622 | 255 | 27 880.55 | 6.92 | |
| StbHLH38 | PGSC0003DMT400063117 | 437 | 48 174.88 | 6.79 | StbHLH76 | PGSC0003DMT400018274 | 224 | 25 831.73 | 6.32 | |
| StbHLH77 | PGSC0003DMT400049504 | 485 | 52 745.68 | 6.07 | StbHLH93 | PGSC0003DMT400016793 | 348 | 36 847.65 | 8.57 | |
| StbHLH78 | PGSC0003DMT400002963 | 598 | 65 258.62 | 5.92 | StbHLH94 | PGSC0003DMT400064399 | 388 | 42 575.81 | 5.83 | |
| StbHLH79 | PGSC0003DMT400015017 | 456 | 51 791.64 | 6.22 | StbHLH95 | PGSC0003DMT400028057 | 389 | 44 514.29 | 5.88 | |
| StbHLH80 | PGSC0003DMT400021165 | 435 | 49 379.65 | 5.89 | StbHLH96 | PGSC0003DMT400008370 | 198 | 22 043.97 | 8.88 | |
| StbHLH81 | PGSC0003DMT400053651 | 319 | 36 510.05 | 4.85 | StbHLH97 | PGSC0003DMT400089728 | 305 | 34 320.40 | 5.48 | |
| StbHLH82 | PGSC0003DMT400095303 | 311 | 35 808.26 | 4.90 | StbHLH98 | PGSC0003DMT400076759 | 180 | 20 751.84 | 9.58 | |
| StbHLH83 | PGSC0003DMT400045210 | 330 | 36 732.34 | 6.10 | StbHLH99 | PGSC0003DMT400021130 | 360 | 39 675.21 | 5.92 | |
| StbHLH84 | PGSC0003DMT400045204 | 694 | 75 939.94 | 5.47 | StbHLH100 | PGSC0003DMT400021062 | 380 | 41 565.47 | 9.10 | |
| StbHLH85 | PGSC0003DMT400032139 | 360 | 40 669.07 | 6.81 | StbHLH101 | PGSC0003DMT400041644 | 237 | 27 276.91 | 5.27 | |
| StbHLH86 | PGSC0003DMT400031899 | 431 | 48 790.06 | 6.71 | StbHLH102 | PGSC0003DMT400073760 | 262 | 30 275.48 | 5.87 | |
| StbHLH87 | PGSC0003DMT400053639 | 492 | 53 992.33 | 7.13 | StbHLH103 | PGSC0003DMT400056716 | 441 | 48 896.06 | 6.65 | |
| StbHLH88 | PGSC0003DMT400048756 | 578 | 62 943.67 | 6.42 | StbHLH104 | PGSC0003DMT400020244 | 382 | 39 561.65 | 6.21 | |
| StbHLH89 | PGSC0003DMT400002512 | 284 | 31 495.30 | 5.46 | StbHLH105 | PGSC0003DMT400013766 | 272 | 31 224.18 | 6.76 | |
| StbHLH90 | PGSC0003DMT400033569 | 590 | 64 760.08 | 5.81 | StbHLH106 | PGSC0003DMT400045820 | 296 | 33 899.60 | 7.02 | |
| StbHLH91 | PGSC0003DMT400076223 | 285 | 32 950.42 | 6.27 | StbHLH107 | PGSC0003DMT400011651 | 413 | 46 698.70 | 5.92 | |
| StbHLH92 | PGSC0003DMT400042791 | 439 | 49 242.61 | 6.06 | StbHLH108 | PGSC0003DMT400061284 | 489 | 54 218.28 | 5.12 |

Fig. 4 Chromosome localization of bHLH family genes in S. tuberosum 1:Root. 2:Leaf. 3:Tuber. 4:Control. 5:Salt. 6:Mannitol. 7:Auxin. 8:Gibberellin. 9:Abscisic acid. 10:Heat. In the figure,red refers to the up regulation of gene expression and blue to the down regulation of gene expression
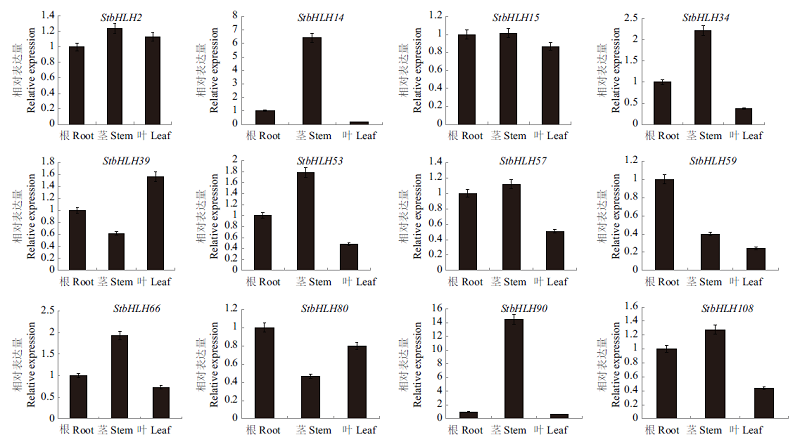
Fig. 6 Tissue expression pattern analysis of bHLH transcription factor in S. tuberosum Abiotic stresses include low potassium,abscisic acid,polyethylene glycol,high salt and hydrogen peroxide. The samples were taken at 0,3,6 and 12 h, respectively
| [1] |
Niu X, Guan Y, Chen S, et al. Genome-wide analysis of basic helix-loop-helix(bHLH)transcription factors in Brachypodium distachyon[J]. BMC Genomics, 2017, 18(1):619.
doi: 10.1186/s12864-017-4044-4 URL |
| [2] | 张全琪, 朱家红, 倪燕妹, 等. 植物bHLH转录因子的结构特点及其生物学功能[J]. 热带亚热带植物学报, 2011, 19(1):84-90. |
| Zhang QQ, Zhu JH, Ni YM, et al. The structure and function of plant bHLH transcription factors[J]. J Trop Subtrop Bot, 2011, 19(1):84-90. | |
| [3] |
Amoutzias GD, Veron AS, Weiner J, et al. One billion years of bZIP transcription factor evolution:conservation and change in dimerization and DNA-binding site specificity[J]. Mol Biol Evol, 2007, 24(3):827-835.
pmid: 17194801 |
| [4] |
Feller A, Machemer K, Braun EL, et al. Evolutionary and comparative analysis of MYB and bHLH plant transcription factors[J]. Plant J, 2011, 66(1):94-116.
doi: 10.1111/tpj.2011.66.issue-1 URL |
| [5] | 杨鹏程, 周波, 李玉花. 植物花青素合成相关的bHLH转录因子[J]. 植物生理学报, 2012, 48(8):747-758. |
|
Yang PC, Zhou B, Li YH. The bHLH transcription factors involved in anthocyanin biosynjournal in plants[J]. Plant Physiol J, 2012, 48(8):747-758.
doi: 10.1104/pp.48.6.747 URL |
|
| [6] | 陈红霖, 胡亮亮, 王丽侠, 等. 绿豆bHLH转录因子家族的鉴定与生物信息学分析[J]. 植物遗传资源学报, 2017, 18(6):1159-1167. |
| Chen HL, Hu LL, Wang LX, et al. Genome-wide identification and bioinformatics analysis of bHLH transcription factor family in mung bean(Vigna radiata L.)[J]. J Plant Genet Resour, 2017, 18(6):1159-1167. | |
| [7] |
Guo XJ, Wang JR. Global identification, structural analysis and expression characterization of bHLH transcription factors in wheat[J]. BMC Plant Biol, 2017, 17(1):90.
doi: 10.1186/s12870-017-1038-y URL |
| [8] |
Song XM, Huang ZN, Duan WK, et al. Genome-wide analysis of the bHLH transcription factor family in Chinese cabbage(Brassica rapa ssp. pekinensis)[J]. Mol Genet Genom, 2014, 289(1):77-91.
doi: 10.1007/s00438-013-0791-3 URL |
| [9] |
Mao K, Dong Q, Li C, et al. Genome wide identification and characterization of apple bHLH transcription factors and expression analysis in response to drought and salt stress[J]. Front Plant Sci, 2017, 8:480.
doi: 10.3389/fpls.2017.00480 pmid: 28443104 |
| [10] | 惠甜, 沈兵琪, 王连春, 等. 桑树bHLH转录因子家族全基因组鉴定与分析[J]. 分子植物育种, 2019, 17(17):5624-5637. |
| Hui T, Shen BQ, Wang LC, et al. Genome-wide identification and analysis of the bHLH transcription factor gene family in Morus notabilis[J]. Mol Plant Breed, 2019, 17(17):5624-5637. | |
| [11] |
Sun W, Jin X, Ma Z, et al. Basic helix-loop-helix(bHLH)gene family in Tartary buckwheat(Fagopyrum tataricum):Genome-wide identification, phylogeny, evolutionary expansion and expression analyses[J]. Int J Biol Macromol, 2020, 155:1478-1490.
doi: 10.1016/j.ijbiomac.2019.11.126 URL |
| [12] |
Tian S, Li L, Wei M, et al. Genome-wide analysis of basic helix-loop-helix superfamily members related to anthocyanin biosynjournal in eggplant(Solanum melongena L.)[J]. PeerJ, 2019, 7:e7768.
doi: 10.7717/peerj.7768 URL |
| [13] | 何昌文, 朱丽, 沈珊, 等. 银杏bHLH91转录因子基因的克隆及表达分析[J]. 广西植物, 2018, 38(2):202-209. |
| He CW, Zhu L, Shen S, et al. Cloning and expression analysis of a bHLH91 transcription factor gene from Ginkgo biloba[J]. Guihaia, 2018, 38(2):202-209. | |
| [14] |
杨梦婷, 张春, 王作平, 等. 玉米ZmbHLH161基因的克隆及功能研究[J]. 作物学报, 2020, 46(12):2008-2016.
doi: 10.3724/SP.J.1006.2020.03022 |
| Yang MT, Zhang C, Wang ZP, et al. Cloning and functional analysis of ZmbHLH161 gene in maize[J]. Acta Agron Sin, 2020, 46(12):2008-2016. | |
| [15] |
Zhang J, Liu B, Li M, et al. The bHLH transcription factor bHLH104 interacts with IAA-LEUCINE RESISTANT3 and modulates iron homeostasis in Arabidopsis[J]. Plant Cell, 2015, 27(3):787-805.
doi: 10.1105/tpc.114.132704 URL |
| [16] |
Liang G, Zhang H, Li X, et al. bHLH transcription factor bHLH115 regulates iron homeostasis in Arabidopsis thaliana[J]. J Exp Bot, 2017, 68(7):1743-1755.
doi: 10.1093/jxb/erx043 pmid: 28369511 |
| [17] |
Li YY, Sui XY, Yang JS, et al. A novel bHLH transcription factor, NtbHLH1, modulates iron homeostasis in tobacco(Nicotiana tabacum L.)[J]. Biochem Biophys Res Commun, 2020, 522(1):233-239.
doi: 10.1016/j.bbrc.2019.11.063 URL |
| [18] |
Yang XM, Ren YL, Cai Y, et al. Overexpression of OsbHLH107, a member of the basic helix-loop-helix transcription factor family, enhances grain size in rice(Oryza sativa L.)[J]. Rice, 2018, 11(1):41.
doi: 10.1186/s12284-018-0237-y URL |
| [19] |
Zhang L, Kang J, Xie Q, et al. The basic helix-loop-helix transcription factor bHLH95 affects fruit ripening and multiple metabolisms in tomato[J]. J Exp Bot, 2020, 71(20):6311-6327.
doi: 10.1093/jxb/eraa363 URL |
| [20] | 何洁, 顾秀容, 魏春华, 等. 西瓜bHLH转录因子家族基因的鉴定及其在非生物胁迫下的表达分析[J]. 园艺学报, 2016, 43(2):281-294. |
| He J, Gu XR, Wei CH, et al. Identification and expression analysis under abiotic stresses of the bHLH transcription factor gene family in watermelon[J]. Acta Hortic Sin, 2016, 43(2):281-294. | |
| [21] |
Le Hir R, Castelain M, Chakraborti D, et al. AtbHLH68 transcription factor contributes to the regulation of ABA homeostasis and drought stress tolerance in Arabidopsis thaliana[J]. Physiol Plantarum, 2017, 160(3):312-327.
doi: 10.1111/ppl.2017.160.issue-3 URL |
| [22] | Chen HC, Cheng WH, Hong CY, et al. The transcription factor OsbHLH035 mediates seed germination and enables seedling recovery from salt stress through ABA-dependent and ABA-independent pathways, respectively[J]. Rice:N Y, 2018, 11(1):50. |
| [23] | 耿晶晶. 甜橙bHLH家族转录因子发掘及CsbHLH18抗寒功能鉴定与作用机制解析[D]. 武汉:华中农业大学, 2018. |
| Geng JJ. Genome-wide identification of bHLH transcription factor family in sweet orange(Citrus sinensis)and functional characterization and mechanism analysis of CsbHLH18 in cold resistance[D]. Wuhan:Huazhong Agricultural University, 2018. | |
| [24] | 刘玉汇. MYB和bHLH转录因子对马铃薯块茎花色素苷生物合成的调控机理研究[D]. 兰州:甘肃农业大学, 2016. |
| Liu YH. Regulatory mechanism of MYB and bHLH transcription factors on anthocyanin biosynthesis in potato Tuber[D]. Lanzhou:Gansu Agricultural University, 2016. | |
| [25] | Tai HH, Goyer C, Murphy AM. Potato MYB and bHLH transcription factors associated with anthocyanin intensity and common scab resistance[J], 2013, 91(10):722-730. |
| [26] |
Wilkins MR, Gasteiger E, Bairoch A, et al. Protein identification and analysis tools in the ExPASy server[J]. Methods Mol Biol, 1999, 112:531-552.
pmid: 10027275 |
| [27] |
Zhang W, Sun ZR. Random local neighbor joining:a new method for reconstructing phylogenetic trees[J]. Mol Phylogenetics Evol, 2008, 47(1):117-128.
doi: 10.1016/j.ympev.2008.01.019 URL |
| [28] |
Potato Genome Sequencing Consortium, Xu X, Pan SK, et al. Genome sequence and analysis of the Tuber crop potato[J]. Nature, 2011, 475(7355):189-195.
doi: 10.1038/nature10158 URL |
| [29] |
Zhou X, Liao Y, Kim SU, et al. Genome-wide identification and characterization of bHLH family genes from Ginkgo biloba[J]. Sci Rep, 2020, 10(1):13723.
doi: 10.1038/s41598-020-69305-3 URL |
| [30] | 王力伟, 房永雨, 刘红葵, 等. bHLH转录因子的研究进展[J]. 畜牧与饲料科学, 2020, 41(1):23-27. |
| Wang LW, Fang YY, Liu HK, et al. Research progress of bHLH transcription factors[J]. Animal Husb Feed Sci, 2020, 41(1):23-27. | |
| [31] | 何开平, 吴楚. bHLH转录因子对植物形态发生的影响[J]. 安徽农业科学, 2010, 38(35):19957-19959. |
| He KP, Wu C. The effects of bHLH transcription factors on plant morphogenesis[J]. J Anhui Agric Sci, 2010, 38(35):19957-19959. | |
| [32] | 王寻, 陈西霞, 李宏亮, 等. 苹果NLP(Nin-like protein)转录因子基因家族全基因组鉴定及表达模式分析[J]. 中国农业科学, 2019, 52(23):4333-4349. |
| Wang X, Chen XX, Li HL, et al. Genome-wide identification and expression pattern analysis of NLP(Nin-like protein)transcription factor gene family in apple[J]. Sci Agric Sin, 2019, 52(23):4333-4349. | |
| [33] | 陈媞颖, 刘娟, 袁媛, 等. 黄芩bHLH转录因子基因家族生物信息学及表达分析[J]. 中草药, 2018, 49(3):671-677. |
| Chen TY, Liu J, Yuan Y, et al. Analysis of bioinformatics and expression level of bHLH transcription factors in Scutellaria baicalensis[J]. Chin Tradit Herb Drugs, 2018, 49(3):671-677. | |
| [34] | 武明珠, 李锋, 王燃, 等. 烟草转录因子bHLH93基因的克隆及表达分析[J]. 烟草科技, 2015, 48(3):1-7. |
| Wu MZ, Li F, Wang R, et al. Cloning and expression of transcription factor gene b HLH93from Nicotiana tabacum[J]. Tob Sci Technol, 2015, 48(3):1-7. | |
| [35] |
Waseem M, Li N, Su D, et al. Overexpression of a basic helix-loop-helix transcription factor gene, SlbHLH22, promotes early flowering and accelerates fruit ripening in tomato(Solanum lycopersicum L.)[J]. Planta, 2019, 250(1):173-185.
doi: 10.1007/s00425-019-03157-8 URL |
| [1] | WANG Zi-ying, LONG Chen-jie, FAN Zhao-yu, ZHANG Lei. Screening of OsCRK5-interacted Proteins in Rice Using Yeast Two-hybrid System [J]. Biotechnology Bulletin, 2023, 39(9): 117-125. |
| [2] | LIU Wen-jin, MA Rui, LIU Sheng-yan, YANG Jiang-wei, ZHANG Ning, SI Huai-jun. Cloning of StCIPK11 Gene and Analysis of Its Response to Drought Stress in Solanum tuberosum [J]. Biotechnology Bulletin, 2023, 39(9): 147-155. |
| [3] | HAN Hao-zhang, ZHANG Li-hua, LI Su-hua, ZHAO Rong, WANG Fang, WANG Xiao-li. Construction of cDNA Library of Cinnamomun bodinieri Induced by Saline-alkali Stress and Screening of CbP5CS Upstream Regulators [J]. Biotechnology Bulletin, 2023, 39(9): 236-245. |
| [4] | KANG Ling-yun, HAN Lu-lu, HAN De-ping, CHEN Jian-sheng, GAN Han-ling, XING Kai, MA You-ji, CUI Kai. Effect of Melatonin on Protecting the Jejunum Mucosal Epithelial Cells from Oxidative Stress Damage [J]. Biotechnology Bulletin, 2023, 39(9): 291-299. |
| [5] | JIANG Run-hai, JIANG Ran-ran, ZHU Cheng-qiang, HOU Xiu-li. Research Progress in Mechanisms of Microbial-enhanced Phytoremediation for Lead-contaminated Soil [J]. Biotechnology Bulletin, 2023, 39(8): 114-125. |
| [6] | LYU Qiu-yu, SUN Pei-yuan, RAN Bin, WANG Jia-rui, CHEN Qing-fu, LI Hong-you. Cloning, Subcellular Localization and Expression Analysis of the Transcription Factor Gene FtbHLH3 in Fagopyrum tataricum [J]. Biotechnology Bulletin, 2023, 39(8): 194-203. |
| [7] | CHEN Xiao, YU Ming-lan, WU Long-kun, ZHENG Xiao-ming, PANG Hong-bo. Research Progress in lncRNA and Their Responses to Low Temperature Stress in Plant [J]. Biotechnology Bulletin, 2023, 39(7): 1-12. |
| [8] | HU Hai-lin, XU Li, LI Xiao-xu, WANG Chen-can, MEI Man, DING Wen-jing, ZHAO Yuan-yuan. Advances in the Regulation of Plant Growth, Development and Stress Physiology by Small Peptide Hormones [J]. Biotechnology Bulletin, 2023, 39(7): 13-25. |
| [9] | WANG Shuai, FENG Yu-mei, BAI Miao, DU Wei-jun, YUE Ai-qin. Functional Analysis of Soybean Gene GmHMGR Responding to Exogenous Hormones and Abiotic Stresses [J]. Biotechnology Bulletin, 2023, 39(7): 131-142. |
| [10] | SUN Ming-hui, WU Qiong, LIU Dan-dan, JIAO Xiao-yu, WANG Wen-jie. Cloning and Expression Analysis of CsTMFs Gene in Tea Plant [J]. Biotechnology Bulletin, 2023, 39(7): 151-159. |
| [11] | WEI Xi-ya, QIN Zhong-wei, LIANG La-mei, LIN Xin-qi, LI Ying-zhi. Mechanism of Melatonin Seed Priming in Improving Salt Tolerance of Capsicum annuum [J]. Biotechnology Bulletin, 2023, 39(7): 160-172. |
| [12] | YU Hui, WANG Jing, LIANG Xin-xin, XIN Ya-ping, ZHOU Jun, ZHAO Hui-jun. Isolation and Functional Verification of Genes Responding to Iron and Cadmium Stresses in Lycium barbarum [J]. Biotechnology Bulletin, 2023, 39(7): 195-205. |
| [13] | ZHANG Bei, REN Fu-sen, ZHAO Yang, GUO Zhi-wei, SUN Qiang, LIU He-juan, ZHEN Jun-qi, WANG Tong-tong, CHENG Xiang-jie. Advances in the Mechanism of Pepper in the Response to Heat Stress [J]. Biotechnology Bulletin, 2023, 39(7): 37-47. |
| [14] | DING Kai-xin, WANG Li-chun, TIAN Guo-kui, WANG Hai-yan, LI Feng-yun, PAN Yang, PANG Ze, SHAN Ying. Research Progress in Uniconazole Alleviating Plant Drought Damage [J]. Biotechnology Bulletin, 2023, 39(6): 1-11. |
| [15] | KONG De-zhen, DUAN Zhen-yu, WANG Gang, ZHANG Xin, XI Lin-qiao. Physiological Characteristics and Transcriptome Analysis of Sorghum bicolor × S. Sudanense Seedlings Under Salt-alkali Stress [J]. Biotechnology Bulletin, 2023, 39(6): 199-207. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||