








Biotechnology Bulletin ›› 2022, Vol. 38 ›› Issue (3): 264-275.doi: 10.13560/j.cnki.biotech.bull.1985.2021-0768
Previous Articles Next Articles
SU Yuan1( ), ZHU Long-jiao1, CAO Ji-juan2, LIU Jian-long3, XU Wen-tao1(
), ZHU Long-jiao1, CAO Ji-juan2, LIU Jian-long3, XU Wen-tao1( )
)
Received:2021-06-17
Online:2022-03-26
Published:2022-04-06
Contact:
XU Wen-tao
E-mail:sy18223736@163.com;xuwentao@cau.edu.cn
SU Yuan, ZHU Long-jiao, CAO Ji-juan, LIU Jian-long, XU Wen-tao. Development of Fluorescence Quantitative Lyophilized Detection Kit Based on Escherichia coli O157∶H7[J]. Biotechnology Bulletin, 2022, 38(3): 264-275.
| 分类 Classification | 冻干保护剂名称 Name of lyoprotectant | 功能 Function |
|---|---|---|
| 糖类 | 海藻糖、乳糖、蔗糖、果糖、麦芽糖 | 稳定剂 |
| 氨基酸类 | 精氨酸、甘氨酸、色氨酸 | 缓冲剂 |
| 盐类 | 磷酸盐(PBS)、醋酸盐、柠檬酸盐和琥珀酸盐 | 缓冲剂 |
| 聚合物类 | 聚乙烯吡咯烷酮(PVP)、聚乙二醇 | 增强效果 |
| 糖脂类 | 牛血清白蛋白(BSA) | 增强效果 |
| 多元醇类 | 甘露醇、甘油、山梨醇 | 填充剂 |
Table 1 Classification of the lyoprotectant[16]
| 分类 Classification | 冻干保护剂名称 Name of lyoprotectant | 功能 Function |
|---|---|---|
| 糖类 | 海藻糖、乳糖、蔗糖、果糖、麦芽糖 | 稳定剂 |
| 氨基酸类 | 精氨酸、甘氨酸、色氨酸 | 缓冲剂 |
| 盐类 | 磷酸盐(PBS)、醋酸盐、柠檬酸盐和琥珀酸盐 | 缓冲剂 |
| 聚合物类 | 聚乙烯吡咯烷酮(PVP)、聚乙二醇 | 增强效果 |
| 糖脂类 | 牛血清白蛋白(BSA) | 增强效果 |
| 多元醇类 | 甘露醇、甘油、山梨醇 | 填充剂 |
| 引物名称 Primer name | 引物序列 Primer sequence(5'-3') |
|---|---|
| 16S rDNA-F | CAACGCGAAGAACCTTACC |
| 16S rDNA-R | CGACAGCCATGCANCACCT |
| E. coli O157:H7-F | CGTCATGGTACGGGTAATGAA |
| E. coli O157:H7-R | TAACGGCTGCCCGATAATG |
| E. coli O157:H7-T | FAM-TGGTCTCAGCAAATCGAGCCACA-TAMRA |
Table 2 Primers' sequences of qPCR
| 引物名称 Primer name | 引物序列 Primer sequence(5'-3') |
|---|---|
| 16S rDNA-F | CAACGCGAAGAACCTTACC |
| 16S rDNA-R | CGACAGCCATGCANCACCT |
| E. coli O157:H7-F | CGTCATGGTACGGGTAATGAA |
| E. coli O157:H7-R | TAACGGCTGCCCGATAATG |
| E. coli O157:H7-T | FAM-TGGTCTCAGCAAATCGAGCCACA-TAMRA |
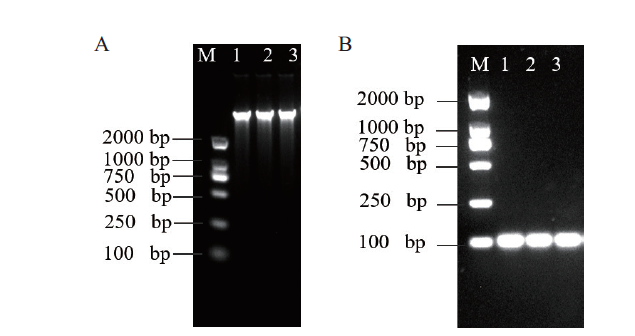
Fig. 1 Agarose gel electrophoresis image A:E. coli O157:H7 genome. M:2 000 bp DNA ladder. 1-3:E. coli O157:H7 template. B:16S rDNA PCR product. M:2 000 bp DNA ladder. 1-3:90 bp target fragment of E. coli O157:H7
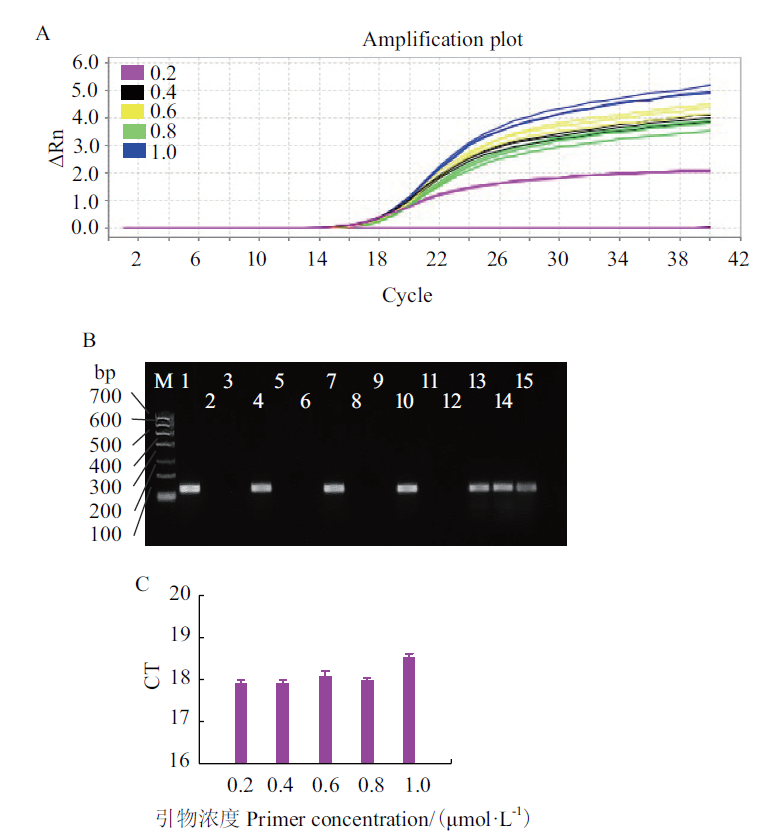
Fig. 2 Concentration optimization of F and R of E. coli O157:H7 A:Quantitative graph. B:Electrophoresis graph. C:Bar graph. M:DNA marker Ι;1-3:0.2 μmol/L F and R,negative control,and control check;4-6:0.3 μmol/L F and R,negative control,and control check;7-9:0.4 μmol/L F and R,negative control,and control check;10-12:0.5 μmol/L F and R,negative control,and control check;13-15:0.6 μmol/L F and R,negative control,and control check
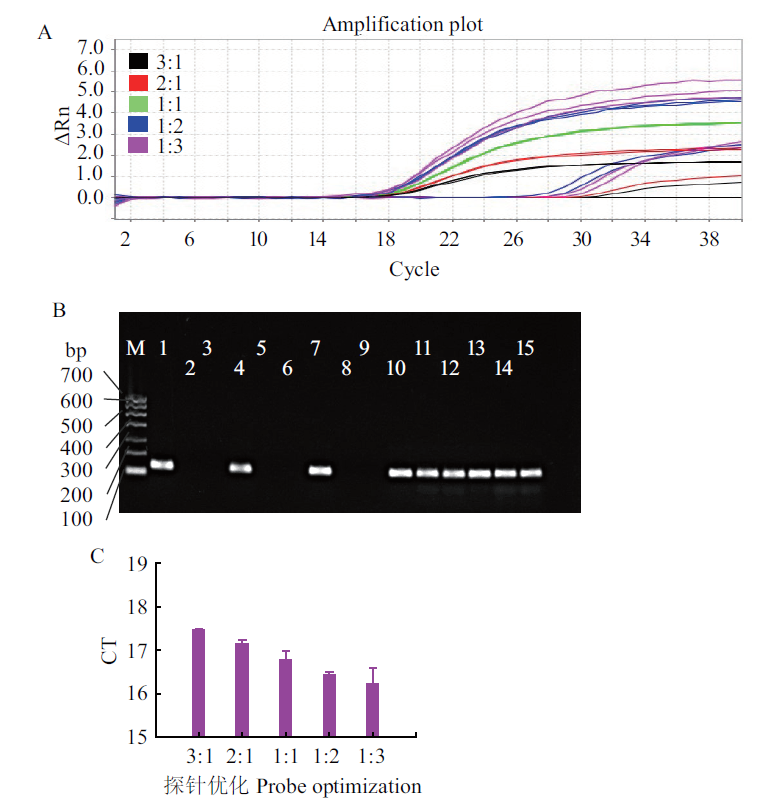
Fig. 3 Optimization of the ratios of primers to probes of E. coli O157:H7 A:Quantitative graph. B:Electrophoresis graph. C:Bar graph. M:DNA marker I;1-3:primer:probe is 3:1,negative control,and control check;4-6:primer:probe is 2:1,negative control,and control check;7-9:primer:probe is 1:1,negative control,and control check;10-12:primer:probe is 1:2,negative control,and control check;13-15:primer:probe is 1:3,negative control,and control check
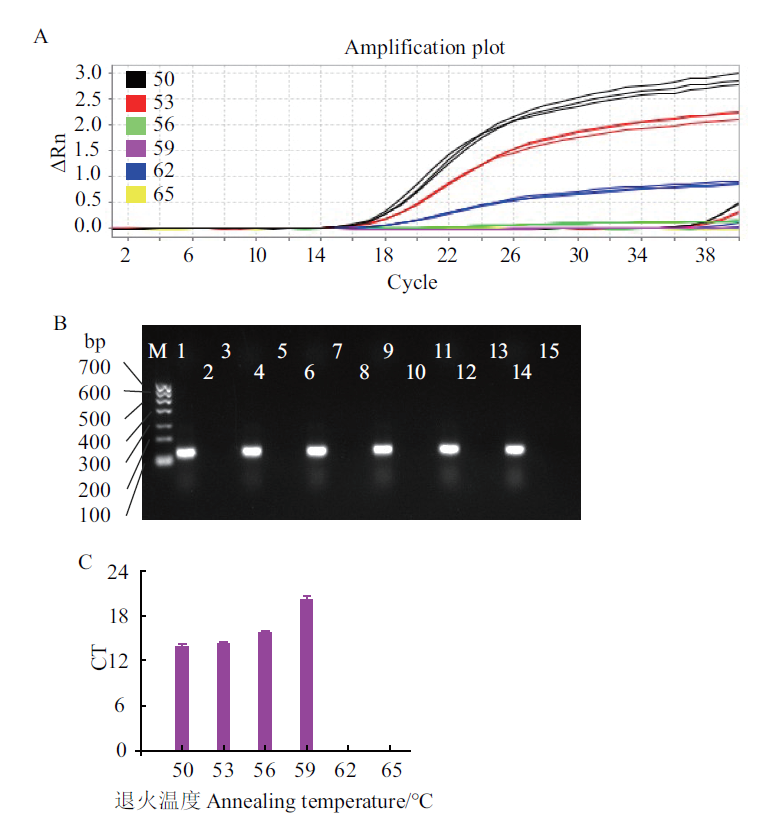
Fig. 4 Optimization of annealing temperature for E. coli O157:H7 A:Quantitative graph. B:Electrophoresis graph. C:Bar graph. M:DNA marker Ι;1-3:50℃,negative control,and control check;4-6:53℃,negative control,control check;7-9:56℃,control check;10-12:59℃,negative control,and control check;13-15:62℃,negative control,and control check;16-18:65℃,negative control,and control check

Fig. 5 Sensitivity of the established E. coli O157:H7 detection method A:Quantitative graph of the sensitivity of the detection method. B:The linear relationship between the Ct and the copies of E. coli O157:H7
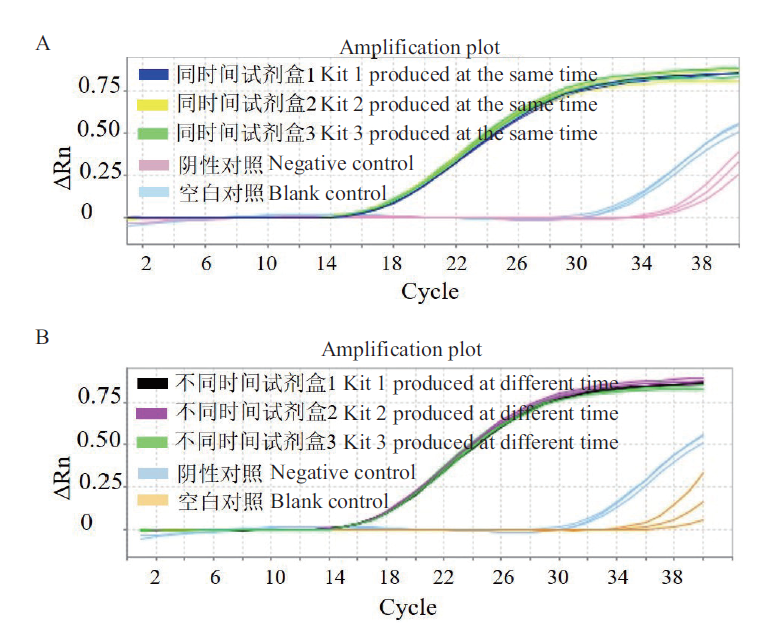
Fig. 10 Reproducibility of the fluorescence quantitative lyophilized detection kit A:Amplification curve of the intra-batch kit. B:Amplification curve of the inter-batch kit
| [1] | Ravenel MP. Food-borne infections and intoxications[J]. JAMA, 1934, 102(13):1106. |
| [2] | 赫鸣睿. 大肠杆菌毒力因子及耐药机制研究进展[J]. 现代畜牧科技, 2018(9):4-5. |
| He MR. Research progress on Escherichia coli virulence factors and drug resistance mechanisms[J]. Mod Animal Husb Sci Technol, 2018(9):4-5. | |
| [3] | 房海. 大肠埃希氏菌[M]. 石家庄: 河北科学技术出版社, 1997. |
| Fang H. Escherichia coli[M]. Shijiazhuang: Hebei Science & Technology Press, 1997. | |
| [4] |
Zhao X, Lin CW, Wang J, et al. Advances in rapid detection methods for foodborne pathogens[J]. J Microbiol Biotechnol, 2014, 24(3):297-312.
doi: 10.4014/jmb.1310.10013 URL |
| [5] |
Lee HM, Yang JS, Yoon SR, et al. Immunomagnetic separation combined with RT-qPCR for evaluating the effect of disinfectant treatments against Norovirus on food contact surfaces[J]. LWT, 2018, 97:83-86.
doi: 10.1016/j.lwt.2018.06.041 URL |
| [6] |
Zhao Y, Zeng D, Yan C, et al. Rapid and accurate detection of Escherichia coli O157∶H7 in beef using microfluidic wax-printed paper-based ELISA[J]. Analyst, 2020, 145(8):3106-3115.
doi: 10.1039/D0AN00224K URL |
| [7] | Mullis KB. The unusual origin of the polymerase chain reaction[J]. Sci Am, 1990,262(4):56- 61,64-5. |
| [8] |
Ibekwe AM, Grieve CM. Detection and quantification of Escherichia coli O157∶H7 in environmental samples by real-time PCR[J]. J Appl Microbiol, 2003, 94(3):421-431.
pmid: 12588551 |
| [9] | 李瑞, 王建昌, 李静, 等. 实时荧光单引物等温扩增(SPIA)技术检测大肠杆菌O157的方法研究[J]. 现代食品科技, 2016, 32(2):317-322, 316. |
| Li R, Wang JC, Li J, et al. A method for the detection of Escherichia coli O157 based on real-time fluorescence single primer isothermal amplification(SPIA)[J]. Mod Food Sci Technol, 2016, 32(2):317-322, 316. | |
| [10] |
Ali AA, Altemimi AB, Alhelfi N, et al. Application of biosensors for detection of pathogenic food bacteria:a review[J]. Biosensors, 2020, 10(6):58.
doi: 10.3390/bios10060058 URL |
| [11] |
Wang WR, Wang XX, Cheng N, et al. Recent advances in nanomaterials-based electrochemical(bio)sensors for pesticides detection[J]. Trac Trends Anal Chem, 2020, 132:116041.
doi: 10.1016/j.trac.2020.116041 URL |
| [12] | Higuchi R, Fockler C, Dollinger G, et al. Kinetic PCR analysis:real-time monitoring of DNA amplification reactions[J]. Biotechnology:N Y, 1993, 11(9):1026-1030. |
| [13] |
Abascal K, Ganora L, Yarnell E. The effect of freeze-drying and its implications for botanical medicine:a review[J]. Phytother Res, 2005, 19(8):655-660.
pmid: 16177965 |
| [14] |
Topuz A, Feng H, Kushad M. The effect of drying method and storage on color characteristics of paprika[J]. LWT Food Sci Technol, 2009, 42(10):1667-1673.
doi: 10.1016/j.lwt.2009.05.014 URL |
| [15] |
Nowak D, Piechucka P, Witrowa-Rajchert D, et al. Impact of material structure on the course of freezing and freeze-drying and on the properties of dried substance, as exemplified by celery[J]. J Food Eng, 2016, 180:22-28.
doi: 10.1016/j.jfoodeng.2016.01.032 URL |
| [16] |
Bjelošević M, Zvonar Pobirk A, Planinšek O, et al. Excipients in freeze-dried biopharmaceuticals:Contributions toward formulation stability and lyophilisation cycle optimisation[J]. Int J Pharm, 2020, 576:119029.
doi: 10.1016/j.ijpharm.2020.119029 URL |
| [17] |
Roos YH. Glass transition temperature and its relevance in food processing[J]. Annu Rev Food Sci Technol, 2010, 1(1):469-496.
doi: 10.1146/food.2010.1.issue-1 URL |
| [18] |
Chi CD, Li XX, Zhang YP, et al. Understanding the effect of freeze-drying on microstructures of starch hydrogels[J]. Food Hydrocoll, 2020, 101:105509.
doi: 10.1016/j.foodhyd.2019.105509 URL |
| [19] |
Sharma VK. Real-time reverse transcription-multiplex PCR for simultaneous and specific detection of rfbE and eae genes of Escherichia coli O157∶H7[J]. Mol Cell Probes, 2006, 20(5):298-306.
doi: 10.1016/j.mcp.2006.03.001 URL |
| [20] | Alsaadi Z H, Tarish A H, Saeed E A. Multiplex PCR rapid and sensitive screening method for detection of local strains of Escherichia coli O157∶H7 in hilla City[J]. Biochemical and Cellular Archives, 2018, 18(1):31-35. |
| [21] | Koev K, Zhelev G, Marutsov P, et al. Molecular screening and characterization of Shiga Toxin-producing Escherichia coli by multiplex PCR assays for stx1, stx2, eaeA, H7 in raw milk[J]. Kafkas Üniversitesi Veteriner Fakültesi Dergisi, 2019, 25(2):271-275. |
| [22] | 王建昌, 王金凤, 段永生, 等. 基于内参的大肠埃希氏菌O157∶H7实时荧光定量PCR快速检测方法的建立[J]. 食品科学, 2015, 36(20):226-231. |
| Wang JC, Wang JF, Duan YS, et al. Development of real-time quantitative PCR assay for the detection of E. coli O157∶H7 based on internal amplification reference[J]. Food Sci, 2015, 36(20):226-231. | |
| [23] | 柯振华. 餐饮及相关食品中病原菌活菌多重实时荧光PCR检测方法的研究与开发[J]. 生物技术进展, 2020, 10(6):717-727. |
| Ke ZH. Research and development of multiplex real-time fluorescence PCR detection method for live pathogenic bacteria in food and beverage[J]. Curr Biotechnol, 2020, 10(6):717-727. | |
| [24] |
Pansare SK, Patel SM. Practical considerations for determination of glass transition temperature of a maximally freeze concentrated solution[J]. AAPS PharmSciTech, 2016, 17(4):805-819.
doi: 10.1208/s12249-016-0551-x URL |
| [25] |
Kasper JC, Winter G, Friess W. Recent advances and further challenges in lyophilization[J]. Eur J Pharm Biopharm, 2013, 85(2):162-169.
doi: 10.1016/j.ejpb.2013.05.019 URL |
| [1] | LI Tian-shun, LI Chen-wei, WANG Jia, ZHU Long-Jiao, XU Wen-tao. Efficient Generation of Secondary Libraries During Functional Nucleic Acids Screening [J]. Biotechnology Bulletin, 2023, 39(3): 116-122. |
| [2] | CAO Ying-fang, ZHAO Xin, LIU Shuang, LI Rui-huan, LIU Na, XU Shi-yong, GAO Fang-rui, MA Hui, LAN Qing-kuo, TAN Jian-xin, WANG Yong. Establishment of Real-time Fluorescent Quantitative PCR Detection Method for Genetically Modified Herbicide-tolerant Soybean GE-J12 [J]. Biotechnology Bulletin, 2022, 38(7): 146-152. |
| [3] | ZHOU Zi-qi, ZHANG Yang-zi, LAN Xin-yue, LIU Yang-er, ZHU Long-jiao, XU Wen-tao. Selection and Application of Light-up Nucleic Acid Aptamers [J]. Biotechnology Bulletin, 2022, 38(5): 240-247. |
| [4] | LIU Ning-ning, WANG Xin-xin, LAN Xin-yue, CHU Hua-shuo, CHEN Xu, CHANG Shi-min, LI Teng-fei, XU Wen-tao. G-Triplex Visualization Nucleic Acid Sensor for the Detection of Tetracycline [J]. Biotechnology Bulletin, 2022, 38(10): 106-114. |
| [5] | HU Xiu-wen, LIU Hua, WANG Yu, TANG Xue-ming, WANG Jin-bin, ZENG Hai-juan, JIANG Wei, LI Hong. Application of CRISPR-Cas System in Nucleic Acid Detection [J]. Biotechnology Bulletin, 2021, 37(9): 266-273. |
| [6] | LI Jia-jun, ZHENG Xiao, SHENG Jie, XU Yao. Novel Coronavirus and Research Progress of Related Clinical Detection Methods [J]. Biotechnology Bulletin, 2021, 37(4): 282-292. |
| [7] | WANG Huan-yu, CHANG Hao-wan, ZHANG Chong-qi, JIN Wei-lin, WEI Fang. Comparison of 5 Methods of Evaluating the Expressions of Chimeric Antigen Receptors [J]. Biotechnology Bulletin, 2021, 37(12): 265-273. |
| [8] | ZHENG Fang-fang, LIN Jun-sheng. Selection and Specificity of Nucleic Acid Aptamers for a Proliferation Inducing Ligand [J]. Biotechnology Bulletin, 2021, 37(10): 196-202. |
| [9] | FANG Shun-yan, SONG Dan, LIU Yan-ping, XU Wen-juan, LIU Jia-yao, HAN Xiang-zhi, LONG Feng. Study on Evanescent Wave Fluorescence Aptasensor for Direct and Rapid Detection of Escherichia coli O157∶H7 [J]. Biotechnology Bulletin, 2020, 36(7): 228-234. |
| [10] | YANG Min, LI Shu-ting, YANG Wen-ping, LI Xiang-yang, XU Wen-tao. Research Progress on Functional Nucleic Acid Biosensors Mediated by DNA/Silver Nanoclusters [J]. Biotechnology Bulletin, 2020, 36(6): 245-254. |
| [11] | WU Ya, XU Zhi-hui, ZHANG Biao, ZHAO Dong-fang, CAO Wen-xin, ZHANG Xing-ping. Research Progress of Nucleic Acid Aptamer Optical Biosensor in Kanamycin Detection [J]. Biotechnology Bulletin, 2020, 36(1): 193-201. |
| [12] | XIAO Bing, LUO Yun-bo, HUANG Kun-lun, ZHANG Yuan, XU Wen-tao. Research Progress in the Quantitative and Unitive Detecting Technologies Based on Functional Nucleic Acid and Labeled Fluorescence [J]. Biotechnology Bulletin, 2019, 35(7): 213-221. |
| [13] | XIE Yin-xia, WANG Wei-ran, CHENG Nan, XU Wen-tao. Research Progress on Electrical Signal Molecules in Electrochemical Functional Nucleic Acids Biosensors [J]. Biotechnology Bulletin, 2019, 35(5): 157-169. |
| [14] | WANG Jiao, ZHANG Shui, ZHANG Jing-rou, SHAO Gui-fang, DENG Ming-hua. Cloning and Expression Analysis of CaCOX3 Gene from Pepper Cytoplasmic Male Sterile Line [J]. Biotechnology Bulletin, 2019, 35(4): 1-6. |
| [15] | XIAO Bing, LIU Bang, LUO Yun-bo, HUANG Kun-lun, ZHANG Yuan, LI Xia-ying, ZHANG Xiu-jie, XU Wen-tao, ZHOU Xiang. Research Progress in Quantitative and Unitive Detecting Technologies of Functional Nucleic Acid and Label-Free Fluorescence [J]. Biotechnology Bulletin, 2019, 35(3): 194-202. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||