








Biotechnology Bulletin ›› 2022, Vol. 38 ›› Issue (3): 9-15.doi: 10.13560/j.cnki.biotech.bull.1985.2021-0802
Previous Articles Next Articles
Received:2021-06-24
Online:2022-03-26
Published:2022-04-06
Contact:
ZHANG Bin
E-mail:zhangbin27104@163.com
ZHANG Bin, YANG Xin-xia. Identification of Key Transcription Factors in Response to Salt Stress in Rice[J]. Biotechnology Bulletin, 2022, 38(3): 9-15.
| 基因IDGene ID | 正向引物Forward primer(5'-3') | 反向引物Reverse primer(5'-3') | 注释Description |
|---|---|---|---|
| Os03g0322900 | CCAAGAACAAGCTGGGCGAG | CTTGAACTCCGTCGCCTTCT | OsLEA3-2[ |
| Os11g0454300 | GCTCCAGCTCAAGCTCGTC | CATGGCATGCTGCTGCTC | RAB21[ |
| Os03g0745000 | CATTGCGCGTCATCGTGAAA | GACCTCGTACGTCTTGCACA | OsHsfA2a |
| Os06g0553100 | CCGTTCGTGTGGAAGACGTA | ACCATAGGTGTTGAGCTGGC | OsHsfC2b |
| Os10g0510000 | AGCTATCGTCCACAGGAA | ACCGGAGCTAATCAGAGT | OsActin |
| Os03g0277300 | ACCATCAAGAACGCCGTCAT | TGAGGACATTCTTCTCGCCG | OsHSP70 |
| Os03g0276500 | ATGATCGGCGTGCAGTACAA | GACGGTGCTTCCGAGATAGG | OsHSP71.1 |
Table 1 Primer sequence for qRT-PCR and gene annotation information
| 基因IDGene ID | 正向引物Forward primer(5'-3') | 反向引物Reverse primer(5'-3') | 注释Description |
|---|---|---|---|
| Os03g0322900 | CCAAGAACAAGCTGGGCGAG | CTTGAACTCCGTCGCCTTCT | OsLEA3-2[ |
| Os11g0454300 | GCTCCAGCTCAAGCTCGTC | CATGGCATGCTGCTGCTC | RAB21[ |
| Os03g0745000 | CATTGCGCGTCATCGTGAAA | GACCTCGTACGTCTTGCACA | OsHsfA2a |
| Os06g0553100 | CCGTTCGTGTGGAAGACGTA | ACCATAGGTGTTGAGCTGGC | OsHsfC2b |
| Os10g0510000 | AGCTATCGTCCACAGGAA | ACCGGAGCTAATCAGAGT | OsActin |
| Os03g0277300 | ACCATCAAGAACGCCGTCAT | TGAGGACATTCTTCTCGCCG | OsHSP70 |
| Os03g0276500 | ATGATCGGCGTGCAGTACAA | GACGGTGCTTCCGAGATAGG | OsHSP71.1 |
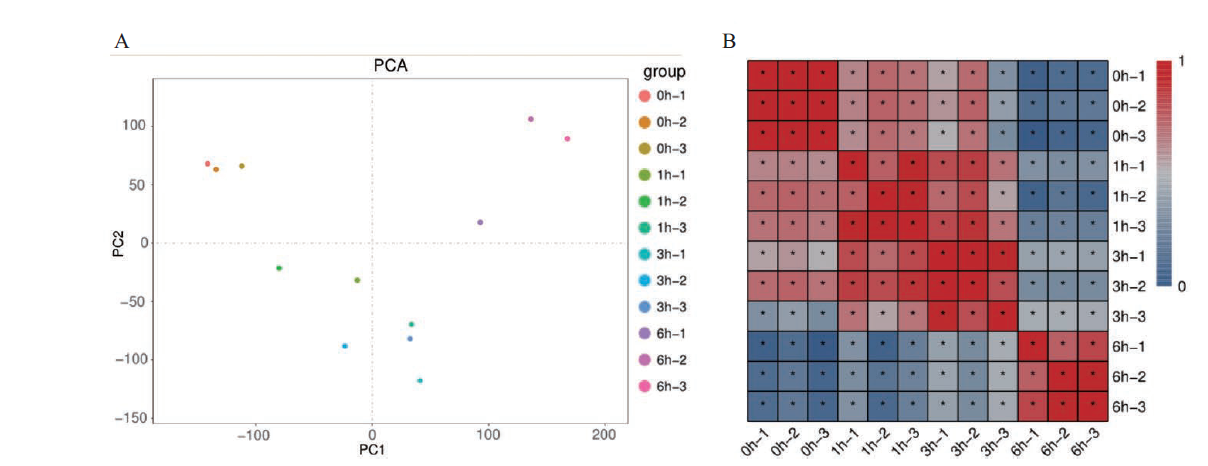
Fig. 1 Relationship among sequencing samples A:PCA analysis diagram of samples. B:Heat map of sample correlation. 0,1,3 and 6 h represent 0,1,3 and 6 h sampling time points. -1,-2 and - 3 represent three repetitions

Fig. 2 Screening of differentially expressed genes(DEGs) and identifical DEGs under salt stress A:Differentially expressed genes under salt stress for 1 h. B:Volcano diagram of differentially expressed genes under salt stress for 3 h. C:Volcano diagram of differentially expressed genes under salt stress for 6 h. D:Venn diagram of differentially expressed genes in response to salt stress
| TF基因TF Gene | STRING中名称STRING ID | 注释Annotation |
|---|---|---|
| Os11g0151002 | OS11T0151002-00 | Os11g0151002蛋白质 Os11g0151002 protein |
| Os06g0553100 | OS06T0553100-01 | 热应激转录因子C-2b Heat stress transcription factor C-2b |
| Os04g0398000 | OS04T0398000-02 | Os04g0398000蛋白 Os04g0398000 protein |
| Os03g0820300 | OsJ_13136 | C2H2转录因子 C2H2 transcription factor |
| Os09g0522100 | DREB1H | 介导高盐和脱水诱导的转录 Mediates high salinity- and dehydration-inducible transcription |
| Os07g0688200 | OS07T0688200-01 | MYB转录因子 MYB transcription factor |
| Os03g0745000 | Hsf4 | 热应激转录因子A-2a Heat stress transcription factor A-2a |
| Os08g0200600 | OS08T0200600-01 | Os08g0200600蛋白质 Os08g0200600 protein |
| Os05g0437100 | OS05T0437100-00 | Os05g0437133蛋白质 Os05g0437133 protein |
| Os12g0564100 | OS12T0564100-01 | 无注释 Annotation not available |
| Os06g0670300 | P0485A07.2 | Os06g0670300蛋白 Os06g0670300 protein |
| Os02g0638650 | P0010C01.6 | 含AP2结构域的类转录因子 AP2 domain-containing transcription factor-like |
| Os01g0693400 | OS01T0693400-01 | 含AP2/ERF和B3结构域的蛋白质 AP2/ERF and B3 domain-containing protein |
| Os11g0168500 | OS11T0168500-00 | 无注释 Annotation not available |
| Os01g0859100 | OS01T0859100-01 | Os01g0859100蛋白 Os01g0859100 protein |
| Os01g0318400 | OS01T0318400-00 | Os01g0318400蛋白质 Os01g0318400 protein |
| Os09g0522200 | ERF24 | 脱水反应元件结合蛋白1A Dehydration-responsive element-binding protein 1A |
| Os03g0820400 | ZFP37 | C2H2型锌指转录因子ZFP37 C2H2 type zinc finger transcription factor ZFP37 |
| Os10g0571600 | OS10T0571600-01 | Os10g0571600蛋白质 Os10g0571600 protein |
| Os01g0626400 | WRKY11 | WRKY转录因子11 WRKY transcription factor 11 |
| Os03g0838800 | OS03T0838800-00 | 锌指蛋白 Zinc finger protein |
| Os05g0132700 | OS05T0132700-01 | Os05g0132700蛋白质 Os05g0132700 protein |
| Os03g0287400 | OsJ_10424 | LOB结构域蛋白1 LOB domain protein 1 |
| Os04g0529100 | OS04T0529100-01 | Os04g0529100蛋白质 Os04g0529100 protein |
| Os11g0154900 | OS11T0154900-01 | BZIP转录因子家族蛋白 BZIP transcription factor family protein |
| Os01g0298400 | OsJ_01424 | P型r2r3MYB蛋白 P-type r2r3 MYB protein |
Table 2 Notes of 26 identical differentially expressed TF genes in STRING
| TF基因TF Gene | STRING中名称STRING ID | 注释Annotation |
|---|---|---|
| Os11g0151002 | OS11T0151002-00 | Os11g0151002蛋白质 Os11g0151002 protein |
| Os06g0553100 | OS06T0553100-01 | 热应激转录因子C-2b Heat stress transcription factor C-2b |
| Os04g0398000 | OS04T0398000-02 | Os04g0398000蛋白 Os04g0398000 protein |
| Os03g0820300 | OsJ_13136 | C2H2转录因子 C2H2 transcription factor |
| Os09g0522100 | DREB1H | 介导高盐和脱水诱导的转录 Mediates high salinity- and dehydration-inducible transcription |
| Os07g0688200 | OS07T0688200-01 | MYB转录因子 MYB transcription factor |
| Os03g0745000 | Hsf4 | 热应激转录因子A-2a Heat stress transcription factor A-2a |
| Os08g0200600 | OS08T0200600-01 | Os08g0200600蛋白质 Os08g0200600 protein |
| Os05g0437100 | OS05T0437100-00 | Os05g0437133蛋白质 Os05g0437133 protein |
| Os12g0564100 | OS12T0564100-01 | 无注释 Annotation not available |
| Os06g0670300 | P0485A07.2 | Os06g0670300蛋白 Os06g0670300 protein |
| Os02g0638650 | P0010C01.6 | 含AP2结构域的类转录因子 AP2 domain-containing transcription factor-like |
| Os01g0693400 | OS01T0693400-01 | 含AP2/ERF和B3结构域的蛋白质 AP2/ERF and B3 domain-containing protein |
| Os11g0168500 | OS11T0168500-00 | 无注释 Annotation not available |
| Os01g0859100 | OS01T0859100-01 | Os01g0859100蛋白 Os01g0859100 protein |
| Os01g0318400 | OS01T0318400-00 | Os01g0318400蛋白质 Os01g0318400 protein |
| Os09g0522200 | ERF24 | 脱水反应元件结合蛋白1A Dehydration-responsive element-binding protein 1A |
| Os03g0820400 | ZFP37 | C2H2型锌指转录因子ZFP37 C2H2 type zinc finger transcription factor ZFP37 |
| Os10g0571600 | OS10T0571600-01 | Os10g0571600蛋白质 Os10g0571600 protein |
| Os01g0626400 | WRKY11 | WRKY转录因子11 WRKY transcription factor 11 |
| Os03g0838800 | OS03T0838800-00 | 锌指蛋白 Zinc finger protein |
| Os05g0132700 | OS05T0132700-01 | Os05g0132700蛋白质 Os05g0132700 protein |
| Os03g0287400 | OsJ_10424 | LOB结构域蛋白1 LOB domain protein 1 |
| Os04g0529100 | OS04T0529100-01 | Os04g0529100蛋白质 Os04g0529100 protein |
| Os11g0154900 | OS11T0154900-01 | BZIP转录因子家族蛋白 BZIP transcription factor family protein |
| Os01g0298400 | OsJ_01424 | P型r2r3MYB蛋白 P-type r2r3 MYB protein |
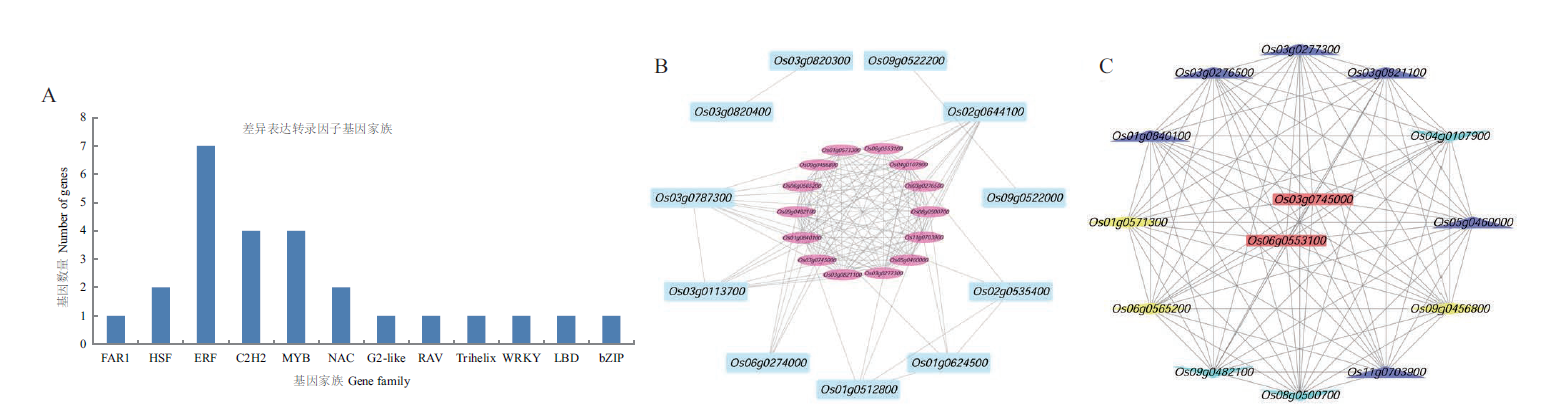
Fig. 3 Classification of the identical differentially expressed genes,important module and key TF screening A:Classification of differentially expressed TF families. B:Screening diagram of important modules. C:Screening map of key TFs:HSP70 gene is shown in the blue triangle. The yellow diamond is HSP90 gene. The green lower triangle is TF gene. The red rectangle is the key Hsf gene
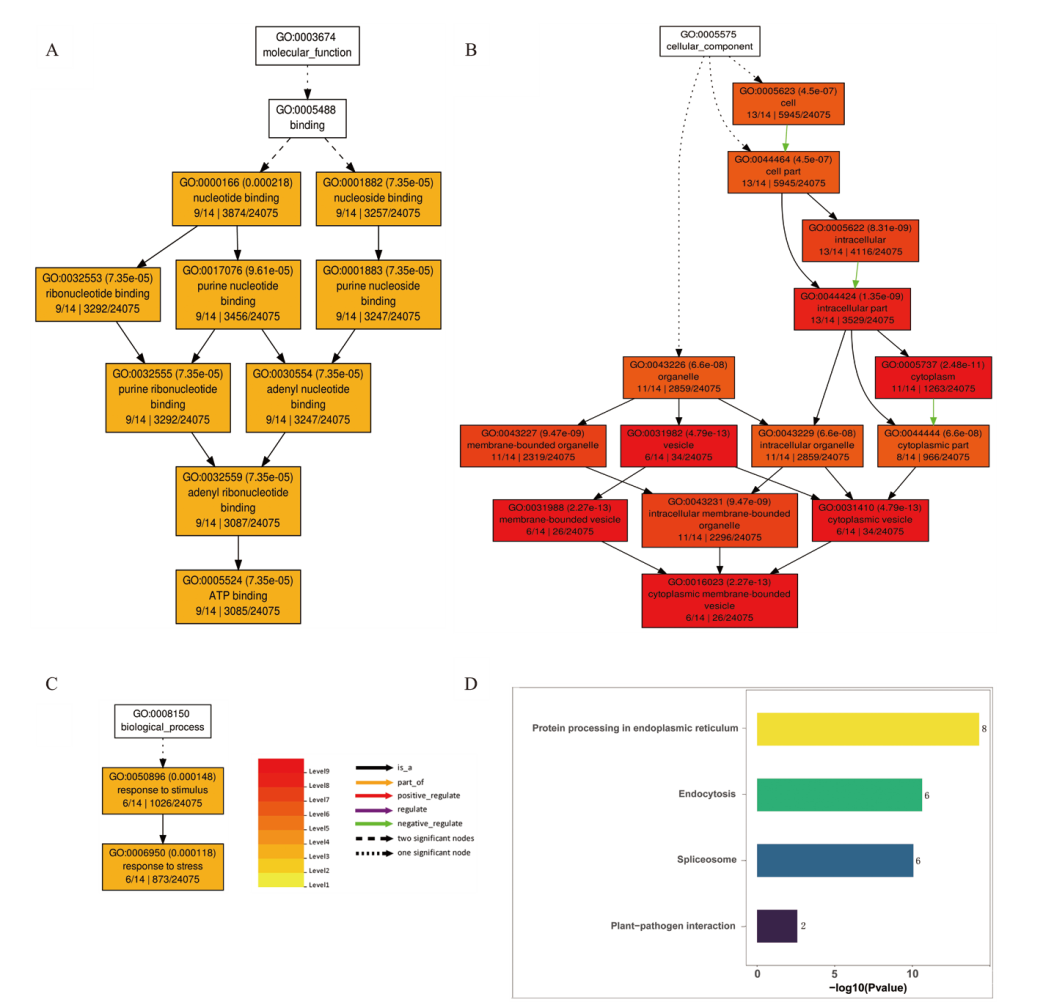
Fig. 4 GO and KEGG enrichment map of important module genes A:GO enrichment analysis of biological function. B:GO enrichment analysis of cell components. C:GO enrichment analysis of biological process. D:KEGG enrichment analysis
| [1] | 朱建峰, 崔振荣, 吴春红, 等. 我国盐碱地绿化研究进展与展望[J]. 世界林业研究, 2018, 31(4):70-75. |
| Zhu JF, Cui ZR, Wu CH, et al. Research advances and prospect of saline and alkali land greening in China[J]. World For Res, 2018, 31(4):70-75. | |
| [2] |
Yang Y, Guo Y. Elucidating the molecular mechanisms mediating plant salt-stress responses[J]. New Phytol, 2018, 217(2):523-539.
doi: 10.1111/nph.14920 URL |
| [3] | 李彬, 王志春, 孙志高, 等. 中国盐碱地资源与可持续利用研究[J]. 干旱地区农业研究, 2005, 23(2):154-158. |
| Li B, Wang ZC, Sun ZG, et al. Resources and sustainable resource exploitation of salinized land in China[J]. Agric Res Arid Areas, 2005, 23(2):154-158. | |
| [4] |
Dai W, Wang M, Gong X, et al. The transcription factor FcWRKY40 of Fortunella crassifolia functions positively in salt tolerance through modulation of ion homeostasis and proline biosynjournal by directly regulating SOS2 and P5CS1 homologs[J]. New Phytol, 2018, 219(3):972-989.
doi: 10.1111/nph.2018.219.issue-3 URL |
| [5] |
Liang W, Ma X, Wan P, et al. Plant salt-tolerance mechanism:a review[J]. Biochem Biophys Res Commun, 2018, 495(1):286-291.
doi: 10.1016/j.bbrc.2017.11.043 URL |
| [6] |
Duan J, Cai W. OsLEA3-2, an abiotic stress induced gene of rice plays a key role in salt and drought tolerance[J]. PLoS One, 2012, 7(9):e45117.
doi: 10.1371/journal.pone.0045117 URL |
| [7] |
Ganguly M, Datta K, Roychoudhury A, et al. Overexpression of Rab16A gene in indica rice variety for generating enhanced salt tolerance[J]. Plant Signal Behav, 2012, 7(4):502-509.
doi: 10.4161/psb.19646 URL |
| [8] | Baillo EH, Kimotho RN, Zhang ZB, et al. Transcription factors associated with abiotic and biotic stress tolerance and their potential for crops improvement[J]. Genes(Basel), 2019, 10(10):771. |
| [9] |
Yamamoto M, Uji S, Sugiyama T, et al. ERdj3B-mediated quality control maintains anther development at high temperatures[J]. Plant Physiol, 2020, 182(4):1979-1990.
doi: 10.1104/pp.19.01356 pmid: 31980572 |
| [10] |
Li X, Zheng H, Shi L, et al. Stress-seventy subfamily A 4, a member of HSP70, confers yeast cadmium tolerance in the loss of mitochondria pyruvate carrier 1[J]. Plant Signal Behav, 2020, 15(2):1719312.
doi: 10.1080/15592324.2020.1719312 URL |
| [11] |
Tiwari LD, Khungar L, Grover A. AtHsc70-1 negatively regulates the basal heat tolerance in Arabidopsis thaliana through affecting the activity of HsfAs and Hsp101[J]. Plant J, 2020, 103(6):2069-2083.
doi: 10.1111/tpj.v103.6 URL |
| [12] |
Biswas S, Islam MN, Sarker S, et al. Overexpression of heterotrimeric G protein beta subunit gene(OsRGB1)confers both heat and salinity stress tolerance in rice[J]. Plant Physiol Biochem, 2019, 144:334-344.
doi: 10.1016/j.plaphy.2019.10.005 URL |
| [13] |
Leng L, Liang Q, Jiang J, et al. A subclass of HSP70s regulate development and abiotic stress responses in Arabidopsis thaliana[J]. J Plant Res, 2017, 130(2):349-363.
doi: 10.1007/s10265-016-0900-6 URL |
| [14] |
Mittal D, Madhyastha DA, Grover A. Gene expression analysis in response to low and high temperature and oxidative stresses in rice:combination of stresses evokes different transcriptional changes as against stresses applied individually[J]. Plant Sci, 2012, 197:102-113.
doi: 10.1016/j.plantsci.2012.09.008 URL |
| [15] |
Chauhan H, Khurana N, Agarwal P, et al. Heat shock factors in rice(Oryza sativa L.):genome-wide expression analysis during reproductive development and abiotic stress[J]. Mol Genet Genom, 2011, 286(2):171.
doi: 10.1007/s00438-011-0638-8 URL |
| [16] |
Mittal D, Enoki Y, Lavania D, et al. Binding affinities and interactions among different heat shock element types and heat shock factors in rice(Oryza sativa L.)[J]. FEBS J, 2011, 278(17):3076-3085.
doi: 10.1111/j.1742-4658.2011.08229.x URL |
| [17] |
Singh A, Mittal D, Lavania D, et al. OsHsfA2c and OsHsfB4b are involved in the transcriptional regulation of cytoplasmic OsClpB(Hsp100)gene in rice(Oryza sativa L.)[J]. Cell Stress Chaperones, 2012, 17(2):243-254.
doi: 10.1007/s12192-011-0303-5 URL |
| [18] |
Yokotani N, Ichikawa T, Kondou Y, et al. Expression of rice heat stress transcription factor OsHsfA2e enhances tolerance to environmental stresses in transgenic Arabidopsis[J]. Planta, 2008, 227(5):957-967.
doi: 10.1007/s00425-007-0670-4 pmid: 18064488 |
| [1] | LIN Hong-yan, GUO Xiao-rui, LIU Di, LI Hui, LU Hai. Molecular Mechanism of Transcriptional Factor AtbHLH68 in Regulating Cell Wall Development by Transcriptome Analysis [J]. Biotechnology Bulletin, 2023, 39(9): 105-116. |
| [2] | WANG Zi-ying, LONG Chen-jie, FAN Zhao-yu, ZHANG Lei. Screening of OsCRK5-interacted Proteins in Rice Using Yeast Two-hybrid System [J]. Biotechnology Bulletin, 2023, 39(9): 117-125. |
| [3] | MIAO Yong-mei, MIAO Cui-ping, YU Qing-cai. Properties of Bacillus subtilis Strain BBs-27 Fermentation Broth and the Inhibition of Lipopeptides Against Fusarium culmorum [J]. Biotechnology Bulletin, 2023, 39(9): 255-267. |
| [4] | WU Yuan-ming, LIN Jia-yi, LIU Yu-xi, LI Dan-ting, ZHANG Zong-qiong, ZHENG Xiao-ming, PANG Hong-bo. Identification of Rice Plant Height-associated QTL Using BSA-seq and RNA-seq [J]. Biotechnology Bulletin, 2023, 39(8): 173-184. |
| [5] | FU Yu, JIA Rui-rui, HE He, WANG Liang-gui, YANG Xiu-lian. Growth Differences Among Grafted Seedlings with Two Rootstocks of Catalpa bungei and Comparative Analysis of Transcriptome [J]. Biotechnology Bulletin, 2023, 39(8): 251-261. |
| [6] | YAO Sha-sha, WANG Jing-jing, WANG Jun-jie, LIANG Wei-hong. Molecular Mechanisms of Rice Grain Size Regulation Related to Plant Hormone Signaling Pathways [J]. Biotechnology Bulletin, 2023, 39(8): 80-90. |
| [7] | WANG Shuai, FENG Yu-mei, BAI Miao, DU Wei-jun, YUE Ai-qin. Functional Analysis of Soybean Gene GmHMGR Responding to Exogenous Hormones and Abiotic Stresses [J]. Biotechnology Bulletin, 2023, 39(7): 131-142. |
| [8] | WEI Xi-ya, QIN Zhong-wei, LIANG La-mei, LIN Xin-qi, LI Ying-zhi. Mechanism of Melatonin Seed Priming in Improving Salt Tolerance of Capsicum annuum [J]. Biotechnology Bulletin, 2023, 39(7): 160-172. |
| [9] | LI Yu, LI Su-zhen, CHEN Ru-mei, LU Hai-qiang. Advances in the Regulation of Iron Homeostasis by bHLH Transcription Factors in Plant [J]. Biotechnology Bulletin, 2023, 39(7): 26-36. |
| [10] | KONG De-zhen, DUAN Zhen-yu, WANG Gang, ZHANG Xin, XI Lin-qiao. Physiological Characteristics and Transcriptome Analysis of Sorghum bicolor × S. Sudanense Seedlings Under Salt-alkali Stress [J]. Biotechnology Bulletin, 2023, 39(6): 199-207. |
| [11] | LIANG Cheng-gang, WANG Yan, LI Tian, OHSUGI Ryu, AOKI Naohiro. Effect of SP1 on Panicle Architecture by Regulating Carbohydrate Remobilization [J]. Biotechnology Bulletin, 2023, 39(5): 152-159. |
| [12] | LIU Hui, LU Yang, YE Xi-miao, ZHOU Shuai, LI Jun, TANG Jian-bo, CHEN En-fa. Comparative Transcriptome Analysis of Cadmium Stress Response Induced by Exogenous Sulfur in Tartary Buckwheat [J]. Biotechnology Bulletin, 2023, 39(5): 177-191. |
| [13] | ZHOU Ding-ding, LI Hui-hu, TANG Xing-yong, YU Fa-xin, KONG Dan-yu, LIU Yi. Research Progress in the Biosynthesis and Regulation of Glycyrrhizic Acid and Liquiritin [J]. Biotechnology Bulletin, 2023, 39(5): 44-53. |
| [14] | WANG Hai-long, LI Yu-qian, WANG Bo, XING Guo-fang, ZHANG Jie-wei. Isolation and Expression Analysis of SiMAPK3 in Setaria italica L. [J]. Biotechnology Bulletin, 2023, 39(3): 123-132. |
| [15] | XIE Yang, XING Yu-meng, ZHOU Guo-yan, LIU Mei-yan, YIN Shan-shan, YAN Li-ying. Transcriptome Analysis of Diploid and Autotetraploid in Cucumber Fruit [J]. Biotechnology Bulletin, 2023, 39(3): 152-162. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
