








Biotechnology Bulletin ›› 2022, Vol. 38 ›› Issue (4): 193-201.doi: 10.13560/j.cnki.biotech.bull.1985.2021-1065
Previous Articles Next Articles
ZHAO Zeng-qiang1,2( ), GUO Wen-ting1, ZHANG Xi1, LI Xiao-ling1, ZHANG Wei1(
), GUO Wen-ting1, ZHANG Xi1, LI Xiao-ling1, ZHANG Wei1( )
)
Received:2021-08-19
Online:2022-04-26
Published:2022-05-06
Contact:
ZHANG Wei
E-mail:tlx4109@126.com;zhw_agr@shzu.edu.cn
ZHAO Zeng-qiang, GUO Wen-ting, ZHANG Xi, LI Xiao-ling, ZHANG Wei. Cloning and Functional Analysis of GhERF5-4D Gene Related to Fusarium oxysporum Resistance in Cotton[J]. Biotechnology Bulletin, 2022, 38(4): 193-201.
| 引物名称Primer name | 引物序列Primer sequence(5'-3') |
|---|---|
| KpnI-GhERF5-4D-F | GGGGTACCATGGCTTCTTTCGGAGA |
| BamHI-GhERF5-4D-R | CGGGATCCCATAACGGCGAGTCCTG |
| qGhERF5-4D-F | AATCCACCCTAAGCCAACGTC |
| qGhERF5-4D-R | CTTTTCTCGTCATGTTCCTCGAT |
| GhUBQ7-F | GAAGACCTACACCAAGCCCAA |
| GhUBQ7-R | CGGACTCTACTCAATCCCCACC |
| XbaI-GhERF5-4D-CO-F | GCTCTAGATCCCACCCACAAGTTTTAACAT |
| KpnI-GhERF5-4D-CO-R | CGGGTACCCTCCTTTTCCCCGACTCTG |
| GhERF1-F | CTTTAGATCACAACTCGCTTC |
| GhERF1-R | TTTTCTTTCGTCGGGCTT |
| GhPR1-F | TTTAGTAAGGTTTTTGACCGACGAA |
| GhPR1-R | GACTCTGTCCATCTTGGTTGTGCTA |
| GhPR2-F | TTCAAGGTTTGCACTCGGAAGA |
| GhPR2-R | CCACCAGCAGCAGAAGTTATCG |
| GhPR4-F | CTTGATATAAGATTGGTTAGCCC |
| GhPR4-R | TTTGCTTTTGTTGTATGTAGTCG |
| GhPR5-F | TTGGCTCTTACTTCCGACCATCT |
| GhPR5-R | GCCGTGATTCATACAGTTATCCTCA |
| GhWRKY70-F | TGGCAACATACGAAGGACAACACA |
| GhWRKY70-R | CTTCGCTCGTCCCGGAAAATATCGC |
| GhNPR1-F | CACGTGGTGCTGTTGTTGTTACTG |
| GhNPR1-R | GCGAATCGTTGCTTTCTTCTTCA |
Table 1 Table of primers
| 引物名称Primer name | 引物序列Primer sequence(5'-3') |
|---|---|
| KpnI-GhERF5-4D-F | GGGGTACCATGGCTTCTTTCGGAGA |
| BamHI-GhERF5-4D-R | CGGGATCCCATAACGGCGAGTCCTG |
| qGhERF5-4D-F | AATCCACCCTAAGCCAACGTC |
| qGhERF5-4D-R | CTTTTCTCGTCATGTTCCTCGAT |
| GhUBQ7-F | GAAGACCTACACCAAGCCCAA |
| GhUBQ7-R | CGGACTCTACTCAATCCCCACC |
| XbaI-GhERF5-4D-CO-F | GCTCTAGATCCCACCCACAAGTTTTAACAT |
| KpnI-GhERF5-4D-CO-R | CGGGTACCCTCCTTTTCCCCGACTCTG |
| GhERF1-F | CTTTAGATCACAACTCGCTTC |
| GhERF1-R | TTTTCTTTCGTCGGGCTT |
| GhPR1-F | TTTAGTAAGGTTTTTGACCGACGAA |
| GhPR1-R | GACTCTGTCCATCTTGGTTGTGCTA |
| GhPR2-F | TTCAAGGTTTGCACTCGGAAGA |
| GhPR2-R | CCACCAGCAGCAGAAGTTATCG |
| GhPR4-F | CTTGATATAAGATTGGTTAGCCC |
| GhPR4-R | TTTGCTTTTGTTGTATGTAGTCG |
| GhPR5-F | TTGGCTCTTACTTCCGACCATCT |
| GhPR5-R | GCCGTGATTCATACAGTTATCCTCA |
| GhWRKY70-F | TGGCAACATACGAAGGACAACACA |
| GhWRKY70-R | CTTCGCTCGTCCCGGAAAATATCGC |
| GhNPR1-F | CACGTGGTGCTGTTGTTGTTACTG |
| GhNPR1-R | GCGAATCGTTGCTTTCTTCTTCA |

Fig.1 PCR amplification and sequence analysis of GhERF5-4 D gene A:The cDNA sequence and the putative amino acid sequence of the GhERF5-4D(The square frame refers to initiation codon ATG and termination codon TAA,* indicates termination codon,and red square frame indicates conserved domain). B:PCR amplification result of GhERF5-4D. C:Enzyme digesting verification of GhERF5-4D. D:Analysis of structure domain of GhERF5-4D protein. E:Sequence analysis of GhERF5-4D gene promoter

Fig.2 Expression pattern of GhERF5-4 D gene after treat by Fusarium oxysporum in cotton A:Zhongmiansuo 12. B:Xinluzao 7. Different letters indicate significant difference at P = 0.05 level. The same below
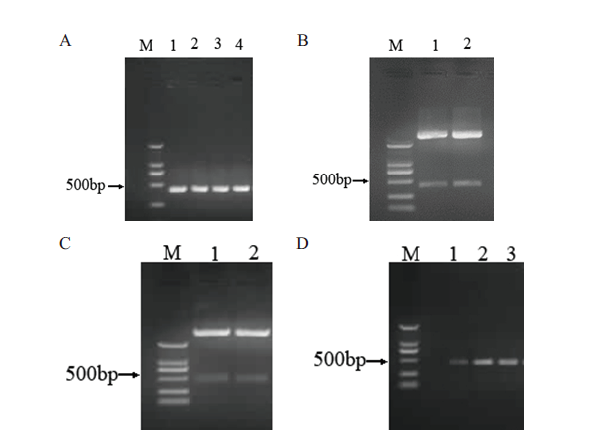
Fig.4 Vector construction of pTRV2-GhERF5-4D-CO A:PCR amplification of GhERF5-4D-CO. B:Enzyme digestion result of GhERF5-4D-CO. C:Enzyme digesting verification of pTRV2-GhERF5-4D-CO. D:PCR amplification result of pTRV2-GhERF5-4D-CD of agrobacterium. M:DL2000
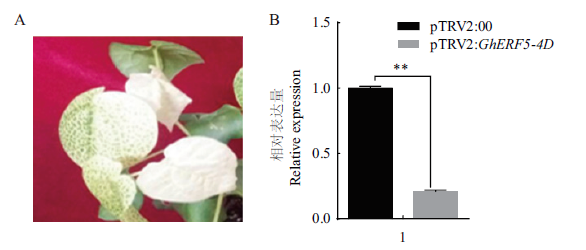
Fig.5 GhERF5-4D gene silencing efficiency and leaf bleaching A:Leaf bleaching after the injection of GhCLA1 gene. B:GhERF5-4D gene silencing efficiency. ** indicates significant difference at P=0.01

Fig.6 GhERF5-4 D gene silencing followed by bacterial treatment A:The incidence phenotype of silencing plants at 12 d. B:The incidence phenot-ype of silencing plants at 20 d. C:20 d stem sections were plated on PDA medium
| [1] | 中华人民共和国统计局. 国家统计局关于2020年棉花产量的公告[EB/OL].(2020-12-18)[2021-12-06]. http://www.stats.gov.cn/tjsj/zxfb/202012/t20201218_1810113.html. |
| [2] | 李菲, 龚记熠, 李欲柯, 等. 抗旱耐盐植物功能基因发掘及其在棉花育种中的应用[J]. 分子植物育种, 2019, 17(22):7395-7400. |
| Li F, Gong JY, Li YK, et al. Excavation of drought and salt responsive functional genes and their application in cotton breeding[J]. Mol Plant Breed, 2019, 17(22):7395-7400. | |
| [3] | 李潇玲, 张析, 郭文婷, 等. 棉花抗枯萎病相关转录因子基因的克隆与表达[J]. 西北植物学报, 2017, 37(1):67-73. |
| Li XL, Zhang X, Guo WT, et al. Cloning and expression of a transcription factor gene related to cotton resistance to Fusarium oxysporum f. sp. vasinfectum[J]. Acta Bot Boreali Occidentalia Sin, 2017, 37(1):67-73. | |
| [4] |
Nakano T, Suzuki K, Fujimura T, et al. Genome-wide analysis of the ERF gene family in Arabidopsis and rice[J]. Plant Physiol, 2006, 140(2):411-432.
doi: 10.1104/pp.105.073783 URL |
| [5] |
Zhuang J, Cai B, Peng RH, et al. Genome-wide analysis of the AP2/ERF gene family in Populus trichocarpa[J]. Biochem Biophys Res Commun, 2008, 371(3):468-474.
doi: 10.1016/j.bbrc.2008.04.087 URL |
| [6] |
Licausi F, Giorgi FM, Zenoni S, et al. Genomic and transcriptomic analysis of the AP2/ERF superfamily in Vitis vinifera[J]. BMC Genomics, 2010, 11:719.
doi: 10.1186/1471-2164-11-719 URL |
| [7] |
Hu L, Liu S. Genome-wide identification and phylogenetic analysis of the ERF gene family in cucumbers[J]. Genet Mol Biol, 2011, 34(4):624-633.
doi: 10.1590/S1415-47572011005000054 URL |
| [8] |
Wessler SR. Homing into the origin of the AP2 DNA binding domain[J]. Trends Plant Sci, 2005, 10(2):54-56.
pmid: 15708341 |
| [9] |
Sakuma Y, Liu Q, Dubouzet JG, et al. DNA-binding specificity of the ERF/AP2 domain of Arabidopsis DREBs, transcription factors involved in dehydration- and cold-inducible gene expression[J]. Biochem Biophys Res Commun, 2002, 290(3):998-1009.
doi: 10.1006/bbrc.2001.6299 URL |
| [10] | 何兰兰. 棉花ERF亚族转录因子基因的克隆与特征分析[D]. 石河子:石河子大学, 2014. |
| He LL. Cloning and characterization analysis of ERF sub-family transcription factor gene from cotton(Gossypium hirsutum L.)[D]. Shihezi:Shihezi University, 2014. | |
| [11] |
Dubouzet JG, Sakuma Y, Ito Y, et al. OsDREBgenes in rice, Oryza sativa L., encode transcription activators that function in drought-, high-salt- and cold-responsive gene expression[J]. Plant J, 2003, 33(4):751-763.
pmid: 12609047 |
| [12] |
Gutterson N, Reuber TL. Regulation of disease resistance pathways by AP2/ERF transcription factors[J]. Curr Opin Plant Biol, 2004, 7(4):465-471.
pmid: 15231271 |
| [13] |
McGrath KC, Dombrecht B, Manners JM, et al. Repressor- and activator-type ethylene response factors functioning in jasmonate signaling and disease resistance identified via a genome-wide screen of Arabidopsis transcription factor gene expression[J]. Plant Physiol, 2005, 139(2):949-959.
pmid: 16183832 |
| [14] |
Berrocal-Lobo M, Molina A, Solano R. Constitutive expression of ETHYLENE-RESPONSE-FACTOR1 in Arabidopsis confers resistance to several necrotrophic fungi[J]. Plant J, 2002, 29(1):23-32.
pmid: 12060224 |
| [15] |
Pré M, Atallah M, Champion A, et al. The AP2/ERF domain transcription factor ORA59 integrates jasmonic acid and ethylene signals in plant defense[J]. Plant Physiol, 2008, 147(3):1347-1357.
doi: 10.1104/pp.108.117523 URL |
| [16] |
Oñate-Sánchez L, Anderson JP, Young J, et al. AtERF14, a member of the ERF family of transcription factors, plays a nonredundant role in plant defense[J]. Plant Physiol, 2007, 143(1):400-409.
doi: 10.1104/pp.106.086637 URL |
| [17] |
Park JM, Park CJ, Lee SB, et al. Overexpression of the tobacco Tsi1 gene encoding an EREBP/AP2-type transcription factor enhances resistance against pathogen attack and osmotic stress in tobacco[J]. Plant Cell, 2001, 13(5):1035-1046.
pmid: 11340180 |
| [18] | Fischer U, Dröge-Laser W. Overexpression of NtERF5, a new member of the tobacco ethylene response transcription factor family enhances resistance to tobacco mosaic virus[J]. Mol Plant Microbe Interactions®, 2004, 17(10):1162-1171. |
| [19] |
Chen X, Guo Z. Tobacco OPBP1 enhances salt tolerance and disease resistance of transgenic rice[J]. Int J Mol Sci, 2008, 9(12):2601-2613.
doi: 10.3390/ijms9122601 URL |
| [20] |
Gu YQ, Wildermuth MC, Chakravarthy S, et al. Tomato transcription factors pti4, pti5, and pti6 activate defense responses when expressed in Arabidopsis[J]. Plant Cell, 2002, 14(4):817-831.
doi: 10.1105/tpc.000794 URL |
| [21] |
Dong N, Liu X, Lu Y, et al. Overexpression of TaPIEP1, a pathogen-induced ERF gene of wheat, confers host-enhanced resistance to fungal pathogen Bipolaris sorokiniana[J]. Funct Integr Genomics, 2010, 10(2):215-226.
doi: 10.1007/s10142-009-0157-4 URL |
| [22] |
Jung J, Won SY, Suh SC, et al. The barley ERF-type transcription factor HvRAF confers enhanced pathogen resistance and salt tolerance in Arabidopsis[J]. Planta, 2007, 225(3):575-588.
doi: 10.1007/s00425-006-0373-2 URL |
| [23] |
Zhang G, Chen M, Li L, et al. Overexpression of the soybean GmERF3 gene, an AP2/ERF type transcription factor for increased tolerances to salt, drought, and diseases in transgenic tobacco[J]. J Exp Bot, 2009, 60(13):3781-3796.
doi: 10.1093/jxb/erp214 URL |
| [24] |
Cao YF, Wu YF, Zheng Z, et al. Overexpression of the rice EREBP-like gene OsBIERF3 enhances disease resistance and salt tolerance in transgenic tobacco[J]. Physiol Mol Plant Pathol, 2005, 67(3/4/5):202-211.
doi: 10.1016/j.pmpp.2006.01.004 URL |
| [25] |
Anderson JP, Lichtenzveig J, Gleason C, et al. The B-3 ethylene response factor MtERF1-1 mediates resistance to a subset of root pathogens in Medicago truncatula without adversely affecting symbiosis with rhizobia[J]. Plant Physiol, 2010, 154(2):861-873.
doi: 10.1104/pp.110.163949 URL |
| [26] |
Jin LG, Liu JY. Molecular cloning, expression profile and promoter analysis of a novel ethylene responsive transcription factor gene GhERF4 from cotton(Gossypium hirstum)[J]. Plant Physiol Biochem, 2008, 46(1):46-53.
doi: 10.1016/j.plaphy.2007.10.004 URL |
| [27] |
Jin LG, Huang B, Li H, et al. Expression profiles and transactivation analysis of a novel ethylene-responsive transcription factor gene GhERF5 from cotton[J]. Prog Nat Sci, 2009, 19(5):563-572.
doi: 10.1016/j.pnsc.2008.05.036 URL |
| [28] | Jin LG, Li H, Liu JY. Molecular characterization of three ethylene responsive element binding factor genes from cotton[J]. J Integr Plant Biol, 2010, 52(5):485-495. |
| [29] |
Zuo KJ, Qin J, Zhao JY, et al. Over-expression GbERF2 transcription factor in tobacco enhances brown spots disease resistance by activating expression of downstream genes[J]. Gene, 2007, 391(1/2):80-90.
doi: 10.1016/j.gene.2006.12.019 URL |
| [30] | 韩泽刚, 赵曾强, 李会会, 等. 枯萎病菌诱导感、抗陆地棉品种的基因表达谱分析[J]. 作物学报, 2015, 41(8):1201-1211. |
|
Han ZG, Zhao ZQ, Li HH, et al. Expression profiling analysis between resistant and susceptible cotton cultivars(Gossypium hirsutum L.)in response to Fusarium wilt[J]. Acta Agron Sin, 2015, 41(8):1201-1211.
doi: 10.3724/SP.J.1006.2015.01201 URL |
|
| [31] | 韩泽刚, 赵曾强, 何兰兰, 等. 枯萎病菌诱导感、抗陆地棉品种的转录因子表达变化[J]. 作物学报, 2015, 41(2):228-239. |
|
Han ZG, Zhao ZQ, He LL, et al. Expression changes of transcription factors in susceptible and resistant upland cotton(Gossypium hirsutum L.)cultivars in response to Fusarium wilt[J]. Acta Agron Sin, 2015, 41(2):228-239.
doi: 10.3724/SP.J.1006.2015.00228 URL |
|
| [32] |
Finatto T, Viana VE, Woyann LG, et al. Can WRKY transcription factors help plants to overcome environmental challenges?[J]. Genet Mol Biol, 2018, 41(3):533-544.
doi: 10.1590/1678-4685-gmb-2017-0232 URL |
| [33] |
Baillo, Kimotho, Zhang, et al. Transcription factors associated with abiotic and biotic stress tolerance and their potential for crops improvement[J]. Genes, 2019, 10(10):771.
doi: 10.3390/genes10100771 URL |
| [34] |
Xie Z, Nolan TM, Jiang H, et al. AP2/ERF transcription factor regulatory networks in hormone and abiotic stress responses in Arabidopsis[J]. Front Plant Sci, 2019, 10:228.
doi: 10.3389/fpls.2019.00228 URL |
| [35] |
Li P, Chai Z, Lin P, et al. Genome-wide identification and expression analysis of AP2/ERF transcription factors in sugarcane(Saccharum spontaneum L.)[J]. BMC Genomics, 2020, 21(1):685.
doi: 10.1186/s12864-020-07076-x URL |
| [36] |
Guo W, Jin L, Miao Y, et al. An ethylene response-related factor, GbERF1-like, from Gossypium barbadense improves resistance to Verticillium dahliae via activating lignin synjournal[J]. Plant Mol Biol, 2016, 91(3):305-318.
doi: 10.1007/s11103-016-0467-6 URL |
| [37] |
Cacas JL, Pré M, Pizot M, et al. GhERF-IIb3 regulates the accumulation of jasmonate and leads to enhanced cotton resistance to blight disease[J]. Mol Plant Pathol, 2017, 18(6):825-836.
doi: 10.1111/mpp.12445 URL |
| [38] |
Caarls L, Van der Does D, Hickman R, et al. Assessing the role of ETHYLENE RESPONSE FACTOR transcriptional repressors in salicylic acid-mediated suppression of jasmonic acid-responsive genes[J]. Plant Cell Physiol, 2017, 58(2):266-278.
doi: 10.1093/pcp/pcw187 pmid: 27837094 |
| [39] |
Champion A, Hebrard E, Parra B, et al. Molecular diversity and gene expression of cotton ERF transcription factors reveal that group IXa members are responsive to jasmonate, ethylene and And[J]. Mol Plant Pathol, 2009, 10(4):471-485.
doi: 10.1111/j.1364-3703.2009.00549.x pmid: 19523101 |
| [40] |
Liu J, Wang Y, Zhao G, et al. A novel Gossypium barbadense ERF transcription factor, GbERFb, regulation host response and resistance to Verticillium dahliae in tobacco[J]. Physiol Mol Biol Plants, 2017, 23(1):125-134.
doi: 10.1007/s12298-016-0402-y URL |
| [41] |
Dong L, Cheng Y, Wu J, et al. Overexpression of GmERF5, a new member of the soybean EAR motif-containing ERF transcription factor, enhances resistance to Phytophthora sojae in soybean[J]. J Exp Bot, 2015, 66(9):2635-2647.
doi: 10.1093/jxb/erv078 URL |
| [42] | 金莉. 棉花GbERF1基因的克隆与功能鉴定[D]. 武汉:华中农业大学, 2012. |
| Jin L. Isolation and characterization of GbERF1 in cotton[D]. Wuhan:Huazhong Agricultural University, 2012. | |
| [43] | Moffat CS, Ingle RA, Wathugala DL, et al. ERF5 and ERF6 play redundant roles as positive regulators of JA/Et-mediated defense against Botrytis cinerea in Arabidopsis[J]. PLoS One, 2012, 7(4):e35995. |
| [44] | 李文正, 张海文, 王俊英, 等. ERF转录因子及其在烟草抗逆性改良中的应用[J]. 生物技术通报, 2006(4):30-34. |
| Li WZ, Zhang HW, Wang JY, et al. Ethylene responsive factors and their application in improving tobacco tolerance to biotic and abiotic stresses[J]. Biotechnol Bull, 2006(4):30-34. |
| [1] | LOU Hui, ZHU Jin-cheng, YANG Yang, ZHANG Wei. Effects of Root Exudates in Resistant and Susceptible Varieties of Cotton on the Growths and Gene Expressions of Fusarium oxysporum [J]. Biotechnology Bulletin, 2023, 39(9): 156-167. |
| [2] | SUN Ming-hui, WU Qiong, LIU Dan-dan, JIAO Xiao-yu, WANG Wen-jie. Cloning and Expression Analysis of CsTMFs Gene in Tea Plant [J]. Biotechnology Bulletin, 2023, 39(7): 151-159. |
| [3] | ZHAO Lin-yan, XU Wu-mei, WANG Hao-ji, WANG Kun-yan, WEI Fu-gang, YANG Shao-zhou, GUAN Hui-lin. Effects of Applying Biochar on the Rhizosphere Fungal Community and Survival Rate of Panax notoginseng Under Continuous Cropping [J]. Biotechnology Bulletin, 2023, 39(7): 219-227. |
| [4] | ZHAO Xue-ting, GAO Li-yan, WANG Jun-gang, SHEN Qing-qing, ZHANG Shu-zhen, LI Fu-sheng. Cloning and Expression of AP2/ERF Transcription Factor Gene ShERF3 in Sugarcane and Subcellular Localization of Its Encoded Protein [J]. Biotechnology Bulletin, 2023, 39(6): 208-216. |
| [5] | YANG Yang, ZHU Jin-cheng, LOU Hui, HAN Ze-gang, ZHANG Wei. Transcriptome Analysis of Interaction Between Gossypium barbadense and Fusarium oxysporum f. sp. vasinfectum [J]. Biotechnology Bulletin, 2023, 39(6): 259-273. |
| [6] | JIANG Qing-chun, DU Jie, WANG Jia-cheng, YU Zhi-he, WANG Yun, LIU Zhong-yu. Expression and Function Analysis of Transcription Factor PcMYB2 from Polygonum cuspidatum [J]. Biotechnology Bulletin, 2023, 39(5): 217-223. |
| [7] | YAO Zi-ting, CAO Xue-ying, XIAO Xue, LI Rui-fang, WEI Xiao-mei, ZOU Cheng-wu, ZHU Gui-ning. Screening of Reference Genes for RT-qPCR in Neoscytalidium dimidiatum [J]. Biotechnology Bulletin, 2023, 39(5): 92-102. |
| [8] | WANG Yi-qing, WANG Tao, WEI Chao-ling, DAI Hao-min, CAO Shi-xian, SUN Wei-jiang, ZENG Wen. Identification and Interaction Analysis of SMAS Gene Family in Tea Plant(Camellia sinensis) [J]. Biotechnology Bulletin, 2023, 39(4): 246-258. |
| [9] | LIU Si-jia, WANG Hao-nan, FU Yu-chen, YAN Wen-xin, HU Zeng-hui, LENG Ping-sheng. Cloning and Functional Analysis of LiCMK Gene in Lilium ‘Siberia’ [J]. Biotechnology Bulletin, 2023, 39(3): 196-205. |
| [10] | WANG Tao, QI Si-yu, WEI Chao-ling, WANG Yi-qing, DAI Hao-min, ZHOU Zhe, CAO Shi-xian, ZENG Wen, SUN Wei-jiang. Expression Analysis and Interaction Protein Validation of CsPPR and CsCPN60-like in Albino Tea Plant(Camellia sinensis) [J]. Biotechnology Bulletin, 2023, 39(3): 218-231. |
| [11] | PANG Qiang-qiang, SUN Xiao-dong, ZHOU Man, CAI Xing-lai, ZHANG Wen, WANG Ya-qiang. Cloning of BrHsfA3 in Chinese Flowering Cabbage and Its Responses to Heat Stress [J]. Biotechnology Bulletin, 2023, 39(2): 107-115. |
| [12] | MIAO Shu-nan, GAO Yu, LI Xin-ru, CAI Gui-ping, ZHANG Fei, XUE Jin-ai, JI Chun-li, LI Run-zhi. Functional Analysis of Soybean GmPDAT1 Genes in the Oil Biosynthesis and Response to Abiotic Stresses [J]. Biotechnology Bulletin, 2023, 39(2): 96-106. |
| [13] | GE Wen-dong, WANG Teng-hui, MA Tian-yi, FAN Zhen-yu, WANG Yu-shu. Genome-wide Identification of the PRX Gene Family in Cabbage(Brassica oleracea L. var. capitata)and Expression Analysis Under Abiotic Stress [J]. Biotechnology Bulletin, 2023, 39(11): 252-260. |
| [14] | YANG Xu-yan, ZHAO Shuang, MA Tian-yi, BAI Yu, WANG Yu-shu. Cloning of Three Cabbage WRKY Genes and Their Expressions in Response to Abiotic Stress [J]. Biotechnology Bulletin, 2023, 39(11): 261-269. |
| [15] | CHEN Chu-yi, YANG Xiao-mei, CHEN Sheng-yan, CHEN Bin, YUE Li-ran. Expression Analysis of the ZF-HD Gene Family in Chrysanthemum nankingense Under Drought and ABA Treatment [J]. Biotechnology Bulletin, 2023, 39(11): 270-282. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||