








Biotechnology Bulletin ›› 2023, Vol. 39 ›› Issue (10): 231-245.doi: 10.13560/j.cnki.biotech.bull.1985.2023-0315
Previous Articles Next Articles
XU Jun1( ), YE Yu-qing1, NIU Ya-jing2, HUANG He1(
), YE Yu-qing1, NIU Ya-jing2, HUANG He1( ), ZHANG Meng-meng2(
), ZHANG Meng-meng2( )
)
Received:2023-04-08
Online:2023-10-26
Published:2023-11-28
Contact:
HUANG He, ZHANG Meng-meng
E-mail:1405549321@qq.com;101navy@163.com;mengmengzhang818@sina.com
XU Jun, YE Yu-qing, NIU Ya-jing, HUANG He, ZHANG Meng-meng. Transcriptome Analysis of Rhizome Development in Chrysanthemum× × morifolium[J]. Biotechnology Bulletin, 2023, 39(10): 231-245.
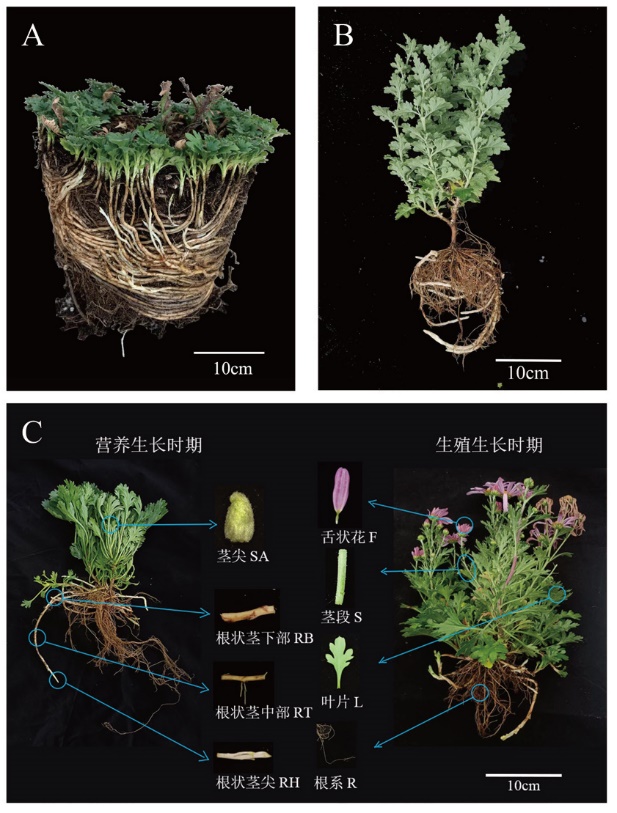
Fig. 1 Schematic diagram of ‘2017XS’ and ‘2005042’ rhizomes and schematic diagram of 8 transcriptome sampling A: Strain with rhizome‘2017XS’. B: Strain with rhizome‘2005042’. C: The material used for transcriptome sequencing is‘2017XS’. The blue circle indicates the specific part of the material, and the arrow indicates the details of the material. Ray florets were taken from the reproductive stage, and other materials were taken from the vegetative stage
| 基因名称Gene name | 基因序列号Gene ID | 正向引物序列Forward primer sequence(5'-3') | 反向引物序列Reverse primer sequence(5'-3') |
|---|---|---|---|
| LHY | c226991.graph_c0 | CGAAGACCGCAGTGCAAATC | TCCCATCCTTTTCTGCCACC |
| BG | c188673.graph_c0 | TGGTCTCGGTTGTTTTGCTTG | TGGAAATGAAGACGGAGTGGAA |
| bZIP | c208538.graph_c0 | GCAATACATATCGGAGCTAGAACG | GCAATCCTTTGCTTGAGGACAC |
| ABA8ox | c209725.graph_c0 | AAGCTCGTTCAAGGCTCGTTAT | TGAGTCCAAGGCGGAGACAG |
| UGT | c177448.graph_c0 | TAACAAGTATGGAAGGAGCAGGTG | GTGGATGGGATGGATGACGA |
| RLP | c217842.graph_c0 | GGATTTCGGGAATGCACTTACT | TCTAATGGAATCGGACCTGCTAAT |
Table 1 Primer sequences for RT-qPCR
| 基因名称Gene name | 基因序列号Gene ID | 正向引物序列Forward primer sequence(5'-3') | 反向引物序列Reverse primer sequence(5'-3') |
|---|---|---|---|
| LHY | c226991.graph_c0 | CGAAGACCGCAGTGCAAATC | TCCCATCCTTTTCTGCCACC |
| BG | c188673.graph_c0 | TGGTCTCGGTTGTTTTGCTTG | TGGAAATGAAGACGGAGTGGAA |
| bZIP | c208538.graph_c0 | GCAATACATATCGGAGCTAGAACG | GCAATCCTTTGCTTGAGGACAC |
| ABA8ox | c209725.graph_c0 | AAGCTCGTTCAAGGCTCGTTAT | TGAGTCCAAGGCGGAGACAG |
| UGT | c177448.graph_c0 | TAACAAGTATGGAAGGAGCAGGTG | GTGGATGGGATGGATGACGA |
| RLP | c217842.graph_c0 | GGATTTCGGGAATGCACTTACT | TCTAATGGAATCGGACCTGCTAAT |
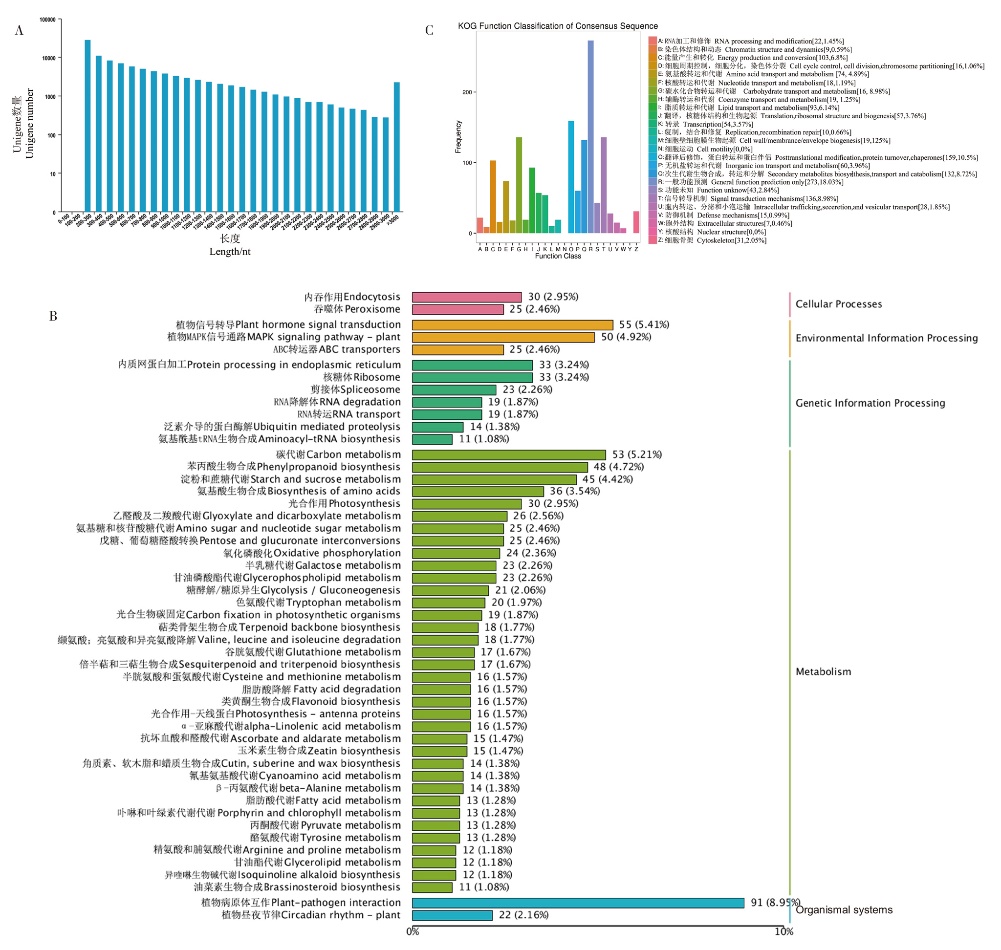
Fig. 2 Unigene length distribution and functional annotation and classification of DEGs in rhizomes A: The distribution of Unigene in different lengths in transcriptome. The X-axis represents the length of Unigene, and the Y-axis represents the number of Unigenes at different lengths. B: Histogram of KOG classification. Unigenes with significant matches in the KOG database are classified into 25 categories. Histograms of different colors represent different KOG categories, and Y-axis represent the number of genes. C: Classification of Unigenes in KEGG pathways. The pink histogram represents the Cellular Process, the yellow histogram represents the Environmental Information Process, the dark green histogram represents the Genetic Information Process, the light green histogram represents the Metabolism, and the blue histogram represents the Organismal system. The number represents the number of genes in this KEGG category and the percentage of the number of genes in this category in the total number of genes
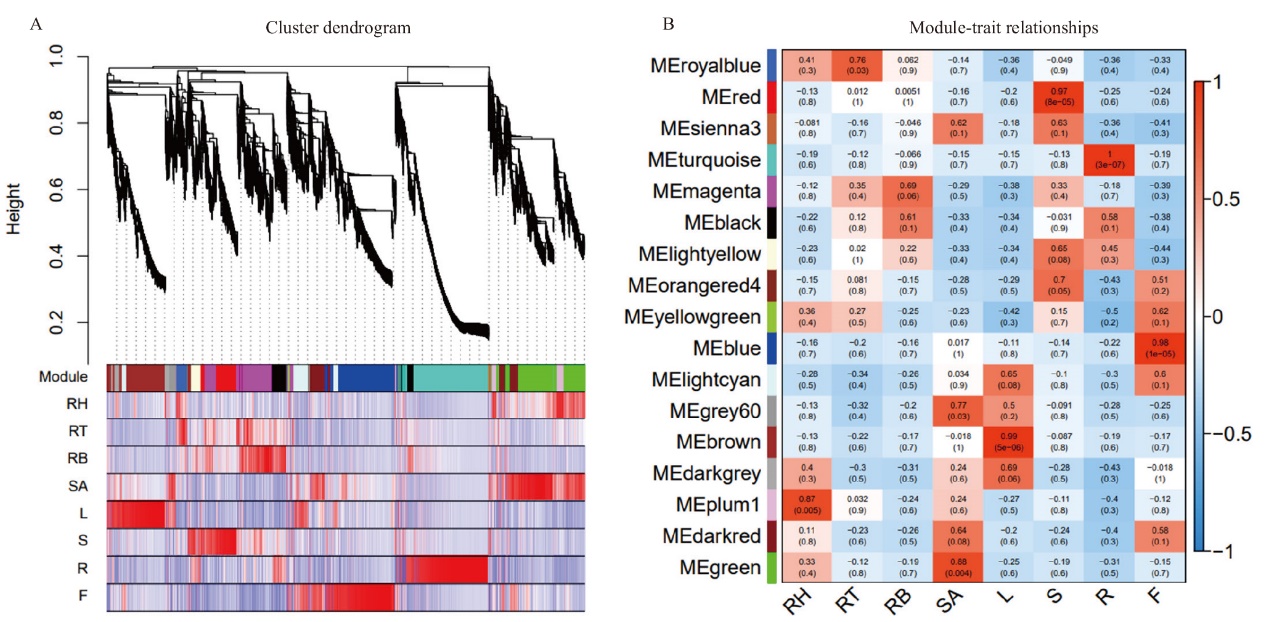
Fig. 3 WGCNA analysis A: Clustering dendrograms of genes and module division. The upper part is the genes cluster dendrograms, the lower part is assigned module, and the same modules had the same color. B: Heat map of the correlation between co-expressed modules and‘2017XS’ tissues. Each row represents a module, and each column represents a tissue. The number in the rectangular box represents the correlation coefficient and corresponding P-value between the module and the trait. Red block represents the positive correlation between the module and the tissues, and blue represents the negative correlation. RH: rhizome apical; RT: middle of rhizome; RB: bottle of rhizome; SA: shoot apical; L: leaf; S: shoot; R: root; F: ray floret
| 差异基因筛选方式 Screening method of DEGs | 基因 Gene | 功能注释 Functional annotation | FPKM | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 根状茎尖 Rhizome tip | 根状茎中部 Rhizome middle | 根状茎下部 Rhizome bottom | 茎尖 Shoot apical | 叶片 Leaf | 茎段 Stem | 根系 Root | 舌状花 Ray floret | |||
| WGCNA | PME68(c195055.graph_c0) | Probable pectinesterase 68(Artemisia annua) | 23.06 | 4.01 | 0.19 | 6.81 | 0.12 | 0.35 | 3.85 | 0.06 |
| PHYB(c200625.graph_c0) | Phytochrome B(Artemisia annua) | 5.83 | 1.45 | 0.57 | 1.18 | 0 | 0.58 | 0 | 0.85 | |
| FLA7(c216061.graph_c0) | Fasciclin-like arabinogalactan protein 7(Tanacetum cinerariifolium) | 71.37 | 7.08 | 0.08 | 14.25 | 4.05 | 9.19 | 0.02 | 19.08 | |
| LEA1(c165828.graph_c1) | Late embryogenesis abundant protein 1(Artemisia annua) | 295.49 | 72.99 | 9.25 | 0.34 | 0.73 | 0 | 0 | 0 | |
| OLE(c190721.graph_c0) | Oleosin(Artemisia annua) | 5.17 | 1.52 | 0.24 | 0.7 | 0.04 | 0.21 | 0 | 0.81 | |
| TIP2-1(c156293.graph_c0) | Aquaporin TIP2-1(Tanacetum cinerariifolium) | 24.91 | 2.7 | 0.29 | 0 | 0 | 0.32 | 0 | 0.32 | |
| LIL3.1(c197767.graph_c1) | Light-harvesting complex-like protein 3 isotype 1(Helianthus annuus) | 7.43 | 0.18 | 0.15 | 0.17 | 0.22 | 0.15 | 0.82 | 0.03 | |
| ABA8ox(c209725.graph_c0) | ABA 8-oxidase(Artemisia annua) | 12.55 | 1.35 | 0.27 | 0.68 | 0.39 | 2.05 | 0.1 | 1.04 | |
| TIP1-1(c185069.graph_c0) | Aquaporin TIP1-1(Artemisia annua) | 503.78 | 74.87 | 6.23 | 51.4 | 32.75 | 21.61 | 0.04 | 6.6 | |
| RD21A(c208052.graph_c0) | Cysteine proteinase COT44(Tanacetum cinerariifolium) | 13.76 | 1.99 | 1.56 | 0.98 | 0.28 | 1.71 | 0.12 | 0.37 | |
| ARP1(c197835.graph_c0) | Probable RNA-binding protein ARP1 isoform X2(Tanacetum cinerariifolium) | 10.34 | 0.74 | 0.07 | 1.05 | 0 | 0.95 | 0 | 0.13 | |
| ARP1(c197835.graph_c1) | Probable RNA-binding protein ARP1 isoform X2(Helianthus annuus) | 50.65 | 6.38 | 0.65 | 4.54 | 0.05 | 5.96 | 0.1 | 0.95 | |
| TIP2-1(c181494.graph_c0) | Aquaporin TIP2-1(Tanacetum cinerariifolium) | 13.52 | 0.95 | 0.16 | 0 | 0 | 0 | 0 | 0 | |
| bZIP34(c208538.graph_c0) | Basic leucine zipper 34(Artemisia annua) | 41.51 | 11.57 | 0.27 | 2.55 | 2.05 | 7.91 | 0 | 2.58 | |
| HIDH(c211020.graph_c0) | 2-hydroxyisoflavanone dehydratase-like(Erigeron canadensis) | 7.48 | 1.18 | 1.16 | 1.96 | 0.08 | 1.97 | 2.16 | 0.03 | |
| VENN | DREB2F(c193648.graph_c0) | Dehydration-responsive element-binding protein 2F(Tanacetum cinerariifolium) | 3.13 | 0.03 | 0.03 | 0.38 | 0 | 0 | 0 | 0 |
| CZF2(c195263.graph_c0) | Zinc finger, CCCH-type(Artemisia annua) | 4.12 | 0.99 | 0.95 | 0.3 | 0.3 | 0.24 | 0.06 | 0.2 | |
| ABA8ox4(c212882.graph_c0) | Abscisic acid 8’-hydroxylase 4(Tanacetum cinerariifolium) | 3.72 | 0.63 | 0.07 | 0.11 | 0.7 | 0.72 | 0.12 | 0.06 | |
| PP2C38(c204872.graph_c0) | Probable protein phosphatase 2C 38(Artemisia annua) | 5.27 | 0.08 | 0.02 | 0.16 | 0.7 | 0 | 0.02 | 0.16 | |
| UGT83A1(c177448.graph_c0) | UDP-glucosyltransferase(Artemisia annua) | 2.8 | 0.06 | 0.1 | 0.25 | 0.23 | 0.05 | 0 | 0 | |
Table 2 Summary of highly expressed genes in the rhizome apical of 2017XS
| 差异基因筛选方式 Screening method of DEGs | 基因 Gene | 功能注释 Functional annotation | FPKM | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 根状茎尖 Rhizome tip | 根状茎中部 Rhizome middle | 根状茎下部 Rhizome bottom | 茎尖 Shoot apical | 叶片 Leaf | 茎段 Stem | 根系 Root | 舌状花 Ray floret | |||
| WGCNA | PME68(c195055.graph_c0) | Probable pectinesterase 68(Artemisia annua) | 23.06 | 4.01 | 0.19 | 6.81 | 0.12 | 0.35 | 3.85 | 0.06 |
| PHYB(c200625.graph_c0) | Phytochrome B(Artemisia annua) | 5.83 | 1.45 | 0.57 | 1.18 | 0 | 0.58 | 0 | 0.85 | |
| FLA7(c216061.graph_c0) | Fasciclin-like arabinogalactan protein 7(Tanacetum cinerariifolium) | 71.37 | 7.08 | 0.08 | 14.25 | 4.05 | 9.19 | 0.02 | 19.08 | |
| LEA1(c165828.graph_c1) | Late embryogenesis abundant protein 1(Artemisia annua) | 295.49 | 72.99 | 9.25 | 0.34 | 0.73 | 0 | 0 | 0 | |
| OLE(c190721.graph_c0) | Oleosin(Artemisia annua) | 5.17 | 1.52 | 0.24 | 0.7 | 0.04 | 0.21 | 0 | 0.81 | |
| TIP2-1(c156293.graph_c0) | Aquaporin TIP2-1(Tanacetum cinerariifolium) | 24.91 | 2.7 | 0.29 | 0 | 0 | 0.32 | 0 | 0.32 | |
| LIL3.1(c197767.graph_c1) | Light-harvesting complex-like protein 3 isotype 1(Helianthus annuus) | 7.43 | 0.18 | 0.15 | 0.17 | 0.22 | 0.15 | 0.82 | 0.03 | |
| ABA8ox(c209725.graph_c0) | ABA 8-oxidase(Artemisia annua) | 12.55 | 1.35 | 0.27 | 0.68 | 0.39 | 2.05 | 0.1 | 1.04 | |
| TIP1-1(c185069.graph_c0) | Aquaporin TIP1-1(Artemisia annua) | 503.78 | 74.87 | 6.23 | 51.4 | 32.75 | 21.61 | 0.04 | 6.6 | |
| RD21A(c208052.graph_c0) | Cysteine proteinase COT44(Tanacetum cinerariifolium) | 13.76 | 1.99 | 1.56 | 0.98 | 0.28 | 1.71 | 0.12 | 0.37 | |
| ARP1(c197835.graph_c0) | Probable RNA-binding protein ARP1 isoform X2(Tanacetum cinerariifolium) | 10.34 | 0.74 | 0.07 | 1.05 | 0 | 0.95 | 0 | 0.13 | |
| ARP1(c197835.graph_c1) | Probable RNA-binding protein ARP1 isoform X2(Helianthus annuus) | 50.65 | 6.38 | 0.65 | 4.54 | 0.05 | 5.96 | 0.1 | 0.95 | |
| TIP2-1(c181494.graph_c0) | Aquaporin TIP2-1(Tanacetum cinerariifolium) | 13.52 | 0.95 | 0.16 | 0 | 0 | 0 | 0 | 0 | |
| bZIP34(c208538.graph_c0) | Basic leucine zipper 34(Artemisia annua) | 41.51 | 11.57 | 0.27 | 2.55 | 2.05 | 7.91 | 0 | 2.58 | |
| HIDH(c211020.graph_c0) | 2-hydroxyisoflavanone dehydratase-like(Erigeron canadensis) | 7.48 | 1.18 | 1.16 | 1.96 | 0.08 | 1.97 | 2.16 | 0.03 | |
| VENN | DREB2F(c193648.graph_c0) | Dehydration-responsive element-binding protein 2F(Tanacetum cinerariifolium) | 3.13 | 0.03 | 0.03 | 0.38 | 0 | 0 | 0 | 0 |
| CZF2(c195263.graph_c0) | Zinc finger, CCCH-type(Artemisia annua) | 4.12 | 0.99 | 0.95 | 0.3 | 0.3 | 0.24 | 0.06 | 0.2 | |
| ABA8ox4(c212882.graph_c0) | Abscisic acid 8’-hydroxylase 4(Tanacetum cinerariifolium) | 3.72 | 0.63 | 0.07 | 0.11 | 0.7 | 0.72 | 0.12 | 0.06 | |
| PP2C38(c204872.graph_c0) | Probable protein phosphatase 2C 38(Artemisia annua) | 5.27 | 0.08 | 0.02 | 0.16 | 0.7 | 0 | 0.02 | 0.16 | |
| UGT83A1(c177448.graph_c0) | UDP-glucosyltransferase(Artemisia annua) | 2.8 | 0.06 | 0.1 | 0.25 | 0.23 | 0.05 | 0 | 0 | |
| 差异基因 筛选方式 Screening method of DEGs | 基因序列号 Gene ID | 功能注释 Functional annotation | FPKM | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 根状茎尖 Rhizome tip | 根状茎中部 Rhizome middle | 根状茎下部 Rhizome bottom | 茎尖 Shoot apical | 叶片 Leaf | 茎段 Stem | 根系 Root | 舌状花 Ray floret | |||
| WGCNA | TFL1(c181499.graph_c0) | TFL1-like protein(Chrysanthemum x morifolium) | 6.19 | 14.30 | 1.24 | 0.14 | 0.20 | 0.07 | 0.17 | 0.04 |
| UVR8(c225307.graph_c0) | Ultraviolet-B receptor UVR8 isoform X2(Cynara cardunculus) | 24.60 | 21.64 | 27.53 | 1.41 | 1.62 | 1.30 | 0.74 | 1.52 | |
| GWD(c220138.graph_c0) | Alpha-glucan water dikinase 1(Helianthus annuus) | 31.29 | 22.66 | 22.05 | 7.28 | 5.81 | 0.10 | 4.84 | 4.22 | |
| UGT73C3(c193961.graph_c0) | UDP-glucosyl transferase 73B2(Lactuca sativa) | 11.45 | 24.51 | 12.62 | 1.14 | 0.49 | 0.65 | 2.62 | 0.32 | |
| LEA1(c165828.graph_c1) | Late embryogenesis abundant protein 1(Artemisia annua) | 295.49 | 72.99 | 9.25 | 0.34 | 0.73 | 0 | 0 | 0 | |
| LEA Dc3(c195245.graph_c0) | Late embryogenesis abundant protein Dc3(Artemisia annua) | 3668.15 | 3341 | 1742.08 | 13.18 | 8.53 | 2.06 | 16.2 | 9.52 | |
| zinc finger(c171393.graph_c0) | Zinc finger, CCHC-type(Artemisia annua) | 1.82 | 15.18 | 3.16 | 0 | 0 | 0.29 | 0.99 | 0.51 | |
| RLP7(c217842.graph_c0) | Leucine-rich repeat protein(Artemisia annua) | 14.87 | 9.42 | 2.48 | 0.02 | 0.07 | 0.09 | 0.13 | 0 | |
| ERF098(c180295.graph_c0) | Ethylene-responsive transcription factor ERF098(Lactuca sativa) | 3.51 | 3.40 | 2.77 | 0.88 | 0.41 | 0 | 0.48 | 0 | |
| PIP1-1(c215772.graph_c0) | Aquaporin PIP1-1-like(Artemisia annua) | 343.98 | 533.32 | 370.29 | 44.36 | 10.71 | 48.83 | 15.59 | 32.56 | |
| hAT transposon(c228735.graph_c0) | Putative hAT transposon superfamily(Tanacetum cinerariifolium) | 895.49 | 1500.66 | 580.71 | 95.56 | 57.82 | 32.38 | 158.83 | 76.66 | |
| GH1(c176119.graph_c0) | Glycoside hydrolase family 1(Artemisia annua) | 12.04 | 27.68 | 15.34 | 0 | 0.13 | 1.02 | 3.03 | 0.73 | |
| SAMDC(c206787.graph_c0) | S-adenosylmethionine decarboxylase proenzyme(Artemisia annua) | 88.53 | 53.73 | 22.24 | 2.34 | 1.76 | 1.26 | 2.14 | 1.16 | |
| LNK1(c223971.graph_c0) | Protein LNK1-like isoform X1(Lactuca sativa) | 14.82 | 14.45 | 16.06 | 1.13 | 0.59 | 1.93 | 0.32 | 0.41 | |
| LHY(c226991.graph_c0) | Late elongated hypocotyl-like(Chrysanthemum seticuspe f. boreale) | 55.07 | 59.39 | 47.02 | 11.46 | 6.01 | 8.76 | 4.67 | 12.26 | |
| TIP2-1(c181494.graph_c0) | Aquaporin TIP2-1(Tanacetum cinerariifolium) | 13.52 | 0.95 | 0.16 | 0 | 0 | 0 | 0 | 0 | |
| RE1(c213815.graph_c1) | Putative RNA-directed DNA polymerase(Helianthus annuus) | 14.31 | 9.52 | 8.36 | 1.19 | 0.32 | 1.76 | 1.61 | 0.38 | |
| BG 46(c188673.graph_c0) | Beta glucosidase 46(Artemisia annua) | 5.00 | 7.08 | 7.03 | 0.15 | 0.80 | 0 | 0.11 | 0 | |
| ACA8(c226840.graph_c0) | Calcium-transporPting ATPase 8(Erigeron canadensis) | 65.4 | 106.47 | 58.25 | 12.76 | 6.78 | 8.67 | 14.69 | 16.91 | |
| DREB1D(c212875.graph_c0) | Dehydration-responsive element-binding factor 1(Artemisia annua) | 13.93 | 8.66 | 2.94 | 0.16 | 0 | 0.04 | 0.25 | 0.13 | |
| MLP43(c91334.graph_c0) | MLP-like protein 43(Tanacetum cinerariifolium) | 128.75 | 232.78 | 41.83 | 8.21 | 0 | 1.35 | 9.98 | 3.82 | |
| K-means | LEA14(c204348.graph_c0) | Late embryogenesis abundant protein(Artemisia annua) | 972.25 | 411.65 | 189.58 | 2.30 | 4.40 | 0.19 | 12.78 | 5.00 |
| ACA10(c210148.graph_c0) | Calcium-transporting ATPase 10(Tanacetum cinerariifolium) | 60.68 | 126.95 | 65.40 | 13.2 | 4.11 | 8.93 | 16.34 | 19.69 | |
| LYK4(c215208.graph_c1) | LysM domain receptor-like kinase 4(Tanacetum cinerariifolium) | 26.77 | 49.34 | 43.47 | 1.80 | 1.66 | 6.83 | 7.13 | 0.44 | |
| PDC2(c187399.graph_c0) | Pyruvate decarboxylase 2(Tanacetum cinerariifolium) | 1.05 | 8.54 | 5.68 | 0 | 0.12 | 0 | 0 | 0 | |
| UGT73(c215576.graph_c0) | UDP-glycosyltransferase 73C7-like(Cynara cardunculus) | 29.51 | 59.45 | 37.95 | 3.22 | 2.8 | 2.83 | 5.55 | 1.07 | |
| zinc finger(c193154.graph_c0) | Zinc finger, CCHC-type(Artemisia annua) | 13.11 | 17.47 | 14.45 | 2.30 | 0.10 | 0.29 | 3.16 | 1.62 | |
| UVR8(c192386.graph_c0) | Ultraviolet-B receptor UVR8-like isoform X1(Erigeron canadensis) | 19.07 | 21.94 | 29.46 | 0.41 | 0.45 | 0.63 | 0 | 0 | |
| 1-SST(c222090.graph_c0) | Sucrose:sucrose 1-fructosyl transferase(Artemisia annua) | 1005.96 | 2853.86 | 1238.04 | 1.80 | 4.62 | 16.83 | 79.72 | 297.35 | |
| MLP43(c195694.graph_c0) | MLP-like protein 43(Artemisia annua) | 2035.38 | 5638.39 | 3866.43 | 224.77 | 17.43 | 77.31 | 669.73 | 75.88 | |
| COL12(c204664.graph_c0) | B-box zinc finger protein 32(Artemisia annua) | 2.50 | 3.98 | 4.90 | 0.14 | 0 | 0 | 0.08 | 0 | |
| VENN | CZF4(c195263.graph_c0) | Zinc finger, CCCH-type(Artemisia annua) | 4.12 | 0.99 | 0.95 | 0.30 | 0.30 | 0.06 | 0.24 | 0.20 |
| ERF C3(c195922.graph_c0) | Ethylene-response factor C3(Helianthus annuus) | 1.13 | 4.14 | 8.48 | 0 | 0 | 0.27 | 0.20 | 0.04 | |
| NDA1(c208704.graph_c0) | NADH dehydrogenase(Artemisia annua) | 1.63 | 1.43 | 3.08 | 0.21 | 0.06 | 0 | 0.46 | 0.27 | |
| BG 46(c196303.graph_c0) | Putative beta-glucosidase(Helianthus annuus) | 1.54 | 3.26 | 1.97 | 0.06 | 0.06 | 0.18 | 0.39 | 0.03 | |
| LEA Dc3(c208154.graph_c0) | Late embryogenesis abundant protein Dc3(Artemisia annua) | 0.19 | 2.36 | 51.39 | 0.05 | 0 | 0.04 | 0 | 0 | |
Table 3 Summary of highly expressed genes in the rhizomes of ‘2017XS’
| 差异基因 筛选方式 Screening method of DEGs | 基因序列号 Gene ID | 功能注释 Functional annotation | FPKM | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 根状茎尖 Rhizome tip | 根状茎中部 Rhizome middle | 根状茎下部 Rhizome bottom | 茎尖 Shoot apical | 叶片 Leaf | 茎段 Stem | 根系 Root | 舌状花 Ray floret | |||
| WGCNA | TFL1(c181499.graph_c0) | TFL1-like protein(Chrysanthemum x morifolium) | 6.19 | 14.30 | 1.24 | 0.14 | 0.20 | 0.07 | 0.17 | 0.04 |
| UVR8(c225307.graph_c0) | Ultraviolet-B receptor UVR8 isoform X2(Cynara cardunculus) | 24.60 | 21.64 | 27.53 | 1.41 | 1.62 | 1.30 | 0.74 | 1.52 | |
| GWD(c220138.graph_c0) | Alpha-glucan water dikinase 1(Helianthus annuus) | 31.29 | 22.66 | 22.05 | 7.28 | 5.81 | 0.10 | 4.84 | 4.22 | |
| UGT73C3(c193961.graph_c0) | UDP-glucosyl transferase 73B2(Lactuca sativa) | 11.45 | 24.51 | 12.62 | 1.14 | 0.49 | 0.65 | 2.62 | 0.32 | |
| LEA1(c165828.graph_c1) | Late embryogenesis abundant protein 1(Artemisia annua) | 295.49 | 72.99 | 9.25 | 0.34 | 0.73 | 0 | 0 | 0 | |
| LEA Dc3(c195245.graph_c0) | Late embryogenesis abundant protein Dc3(Artemisia annua) | 3668.15 | 3341 | 1742.08 | 13.18 | 8.53 | 2.06 | 16.2 | 9.52 | |
| zinc finger(c171393.graph_c0) | Zinc finger, CCHC-type(Artemisia annua) | 1.82 | 15.18 | 3.16 | 0 | 0 | 0.29 | 0.99 | 0.51 | |
| RLP7(c217842.graph_c0) | Leucine-rich repeat protein(Artemisia annua) | 14.87 | 9.42 | 2.48 | 0.02 | 0.07 | 0.09 | 0.13 | 0 | |
| ERF098(c180295.graph_c0) | Ethylene-responsive transcription factor ERF098(Lactuca sativa) | 3.51 | 3.40 | 2.77 | 0.88 | 0.41 | 0 | 0.48 | 0 | |
| PIP1-1(c215772.graph_c0) | Aquaporin PIP1-1-like(Artemisia annua) | 343.98 | 533.32 | 370.29 | 44.36 | 10.71 | 48.83 | 15.59 | 32.56 | |
| hAT transposon(c228735.graph_c0) | Putative hAT transposon superfamily(Tanacetum cinerariifolium) | 895.49 | 1500.66 | 580.71 | 95.56 | 57.82 | 32.38 | 158.83 | 76.66 | |
| GH1(c176119.graph_c0) | Glycoside hydrolase family 1(Artemisia annua) | 12.04 | 27.68 | 15.34 | 0 | 0.13 | 1.02 | 3.03 | 0.73 | |
| SAMDC(c206787.graph_c0) | S-adenosylmethionine decarboxylase proenzyme(Artemisia annua) | 88.53 | 53.73 | 22.24 | 2.34 | 1.76 | 1.26 | 2.14 | 1.16 | |
| LNK1(c223971.graph_c0) | Protein LNK1-like isoform X1(Lactuca sativa) | 14.82 | 14.45 | 16.06 | 1.13 | 0.59 | 1.93 | 0.32 | 0.41 | |
| LHY(c226991.graph_c0) | Late elongated hypocotyl-like(Chrysanthemum seticuspe f. boreale) | 55.07 | 59.39 | 47.02 | 11.46 | 6.01 | 8.76 | 4.67 | 12.26 | |
| TIP2-1(c181494.graph_c0) | Aquaporin TIP2-1(Tanacetum cinerariifolium) | 13.52 | 0.95 | 0.16 | 0 | 0 | 0 | 0 | 0 | |
| RE1(c213815.graph_c1) | Putative RNA-directed DNA polymerase(Helianthus annuus) | 14.31 | 9.52 | 8.36 | 1.19 | 0.32 | 1.76 | 1.61 | 0.38 | |
| BG 46(c188673.graph_c0) | Beta glucosidase 46(Artemisia annua) | 5.00 | 7.08 | 7.03 | 0.15 | 0.80 | 0 | 0.11 | 0 | |
| ACA8(c226840.graph_c0) | Calcium-transporPting ATPase 8(Erigeron canadensis) | 65.4 | 106.47 | 58.25 | 12.76 | 6.78 | 8.67 | 14.69 | 16.91 | |
| DREB1D(c212875.graph_c0) | Dehydration-responsive element-binding factor 1(Artemisia annua) | 13.93 | 8.66 | 2.94 | 0.16 | 0 | 0.04 | 0.25 | 0.13 | |
| MLP43(c91334.graph_c0) | MLP-like protein 43(Tanacetum cinerariifolium) | 128.75 | 232.78 | 41.83 | 8.21 | 0 | 1.35 | 9.98 | 3.82 | |
| K-means | LEA14(c204348.graph_c0) | Late embryogenesis abundant protein(Artemisia annua) | 972.25 | 411.65 | 189.58 | 2.30 | 4.40 | 0.19 | 12.78 | 5.00 |
| ACA10(c210148.graph_c0) | Calcium-transporting ATPase 10(Tanacetum cinerariifolium) | 60.68 | 126.95 | 65.40 | 13.2 | 4.11 | 8.93 | 16.34 | 19.69 | |
| LYK4(c215208.graph_c1) | LysM domain receptor-like kinase 4(Tanacetum cinerariifolium) | 26.77 | 49.34 | 43.47 | 1.80 | 1.66 | 6.83 | 7.13 | 0.44 | |
| PDC2(c187399.graph_c0) | Pyruvate decarboxylase 2(Tanacetum cinerariifolium) | 1.05 | 8.54 | 5.68 | 0 | 0.12 | 0 | 0 | 0 | |
| UGT73(c215576.graph_c0) | UDP-glycosyltransferase 73C7-like(Cynara cardunculus) | 29.51 | 59.45 | 37.95 | 3.22 | 2.8 | 2.83 | 5.55 | 1.07 | |
| zinc finger(c193154.graph_c0) | Zinc finger, CCHC-type(Artemisia annua) | 13.11 | 17.47 | 14.45 | 2.30 | 0.10 | 0.29 | 3.16 | 1.62 | |
| UVR8(c192386.graph_c0) | Ultraviolet-B receptor UVR8-like isoform X1(Erigeron canadensis) | 19.07 | 21.94 | 29.46 | 0.41 | 0.45 | 0.63 | 0 | 0 | |
| 1-SST(c222090.graph_c0) | Sucrose:sucrose 1-fructosyl transferase(Artemisia annua) | 1005.96 | 2853.86 | 1238.04 | 1.80 | 4.62 | 16.83 | 79.72 | 297.35 | |
| MLP43(c195694.graph_c0) | MLP-like protein 43(Artemisia annua) | 2035.38 | 5638.39 | 3866.43 | 224.77 | 17.43 | 77.31 | 669.73 | 75.88 | |
| COL12(c204664.graph_c0) | B-box zinc finger protein 32(Artemisia annua) | 2.50 | 3.98 | 4.90 | 0.14 | 0 | 0 | 0.08 | 0 | |
| VENN | CZF4(c195263.graph_c0) | Zinc finger, CCCH-type(Artemisia annua) | 4.12 | 0.99 | 0.95 | 0.30 | 0.30 | 0.06 | 0.24 | 0.20 |
| ERF C3(c195922.graph_c0) | Ethylene-response factor C3(Helianthus annuus) | 1.13 | 4.14 | 8.48 | 0 | 0 | 0.27 | 0.20 | 0.04 | |
| NDA1(c208704.graph_c0) | NADH dehydrogenase(Artemisia annua) | 1.63 | 1.43 | 3.08 | 0.21 | 0.06 | 0 | 0.46 | 0.27 | |
| BG 46(c196303.graph_c0) | Putative beta-glucosidase(Helianthus annuus) | 1.54 | 3.26 | 1.97 | 0.06 | 0.06 | 0.18 | 0.39 | 0.03 | |
| LEA Dc3(c208154.graph_c0) | Late embryogenesis abundant protein Dc3(Artemisia annua) | 0.19 | 2.36 | 51.39 | 0.05 | 0 | 0.04 | 0 | 0 | |

Fig. 4 Venn diagram of DEGs in three parts of rhizome and other tissues A: DEGs between rhizome tip and non-rhizome tissues. B: DEGs between middle of rhizome and non-rhizome tissues. C: DEGs between bottle of rhizome and non-rhizome tissues. Sets of different colors represent different comparison groups, and numbers represent the number of genes contained in the region. The red box indicates the genes differentially expressed in the five comparison groups. RH: rhizome apical; RT: middle of rhizome; RB: bottle of rhizome; SA: shoot apical; L: leaf; S: shoot; R: root; F: ray floret

Fig. 5 Cluster analysis of gene expression patterns of rhizome stable line ‘2017XS’ The X-axis of each cluster represents the material sequencing organization of the transcriptome, the Y-axis represents the gene expression level in the cluster, and the curve represents the change trend of gene expression level in the cluster. RH: rhizome apical; RT: middle of rhizome; RB: bottle of rhizome; SA: shoot apical; L: leaf; S: shoot; R: root; F: ray floret
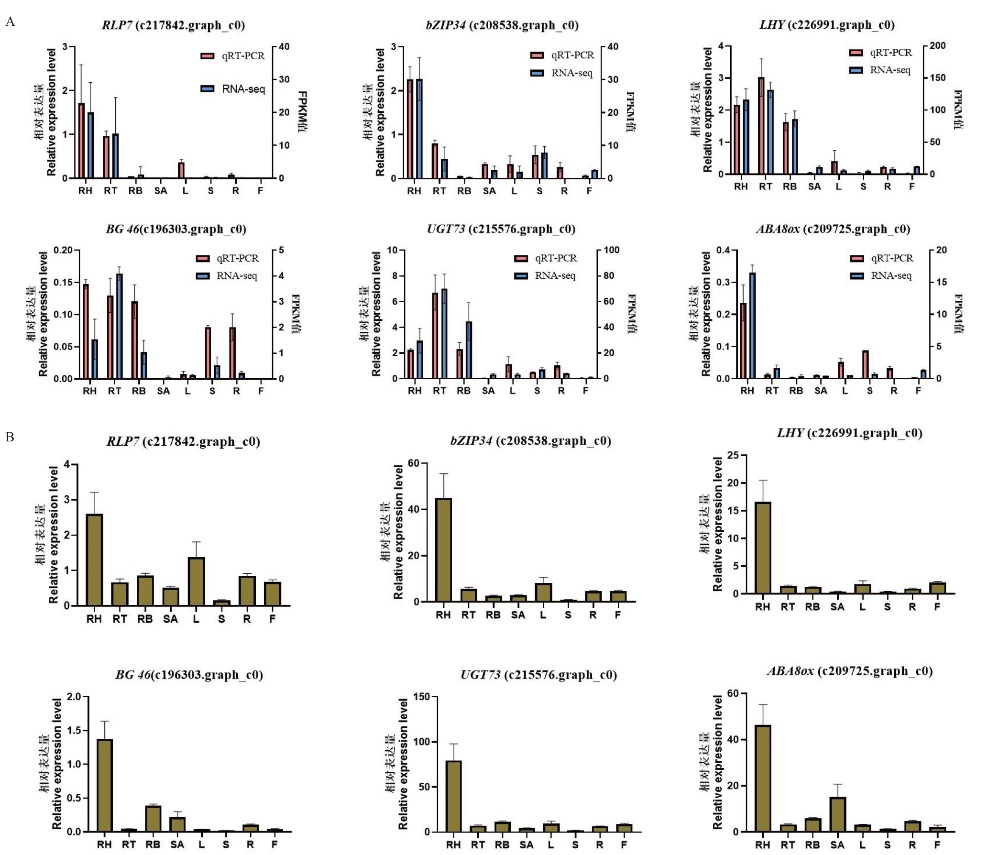
Fig. 6 Verification of transcriptome sequencing data A: RT-qPCR verification of transcriptome-screened genes in ‘2017XS’. B: RT-qPCR verification of transcriptome-screened genes in‘2005042’. RH: rhizome apical; RT: middle of rhizome; RB: bottle of rhizome; SA: shoot apical; L: leaf; S: shoot; R: root; F: ray floret
| [1] |
Yang W, Liu WH, Niu KJ, et al. Transcriptional regulation of different rhizome parts reveal the candidate genes that regulate rhizome development in Poa pratensis[J]. DNA Cell Biol, 2022, 41(2): 151-168.
doi: 10.1089/dna.2021.0337 URL |
| [2] |
Yoshida A, Terada Y, Toriba T, et al. Analysis of rhizome development in Oryza longistaminata, a wild rice species[J]. Plant Cell Physiol, 2016, 57(10): 2213-2220.
pmid: 27516415 |
| [3] |
Ma XQ, Yu JJ, Zhuang LL, et al. Differential regulatory pathways associated with drought-inhibition and post-drought recuperation of rhizome development in perennial grass[J]. Ann Bot, 2020, 126(3): 481-497.
doi: 10.1093/aob/mcaa099 URL |
| [4] |
Zhang SL, Huang GF, Zhang YJ, et al. Sustained productivity and agronomic potential of perennial rice[J]. Nat Sustain, 2023, 6(1): 28-38.
doi: 10.1038/s41893-022-00997-3 |
| [5] |
Anderson N, Gesick E. Phenotypic markers for selection of winter hardy garden chrysanthemum(Dendranthema × grandiflora Tzvelv.) genotypes[J]. Sci Hortic, 2004, 101(1/2): 153-167.
doi: 10.1016/j.scienta.2003.10.006 URL |
| [6] |
Zhang LL, Xu YJ, Liu XN, et al. The chrysanthemum DEAD-box RNA helicase CmRH56 regulates rhizome outgrowth in response to drought stress[J]. J Exp Bot, 2022, 73(16): 5671-5681.
doi: 10.1093/jxb/erac213 URL |
| [7] |
Fu JX, Yang LW, Dai SL. Conservation of Arabidopsis thaliana circadian clock genes in Chrysanthemum lavandulifolium[J]. Plant Physiol Biochem, 2014, 80: 337-347.
doi: 10.1016/j.plaphy.2014.04.001 URL |
| [8] |
Fu JX, Yang LW, Dai SL. Identification and characterization of the CONSTANS-like gene family in the short-day plant Chrysanthemum lavandulifolium[J]. Mol Genet Genomics, 2015, 290(3): 1039-1054.
doi: 10.1007/s00438-014-0977-3 URL |
| [9] |
Hong Y, Yang LW, Li ML, et al. Comparative analyses of light-induced anthocyanin accumulation and gene expression between the ray florets and leaves in chrysanthemum[J]. Plant Physiol Biochem, 2016, 103: 120-132.
doi: 10.1016/j.plaphy.2016.03.006 URL |
| [10] |
Yang M, Zhu LP, Pan C, et al. Transcriptomic analysis of the regulation of rhizome formation in temperate and tropical lotus(Nelumbo nucifera)[J]. Sci Rep, 2015, 5: 13059.
doi: 10.1038/srep13059 pmid: 26279185 |
| [11] |
Cheng LB, Li SY, Yin JJ, et al. Genome-wide analysis of differentially expressed genes relevant to rhizome formation in lotus root(Nelumbo nucifera gaertn)[J]. PLoS One, 2013, 8(6): e67116.
doi: 10.1371/journal.pone.0067116 URL |
| [12] |
Zhang T, Zhao XQ, Wang WS, et al. Deep transcriptome sequencing of rhizome and aerial-shoot in Sorghum propinquum[J]. Plant Mol Biol, 2014, 84(3): 315-327.
doi: 10.1007/s11103-013-0135-z pmid: 24104862 |
| [13] |
Wang KH, Peng HZ, Lin EP, et al. Identification of genes related to the development of bamboo rhizome bud[J]. J Exp Bot, 2010, 61(2): 551-561.
doi: 10.1093/jxb/erp334 pmid: 19965904 |
| [14] |
Grabherr MG, Haas BJ, Yassour M, et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome[J]. Nat Biotechnol, 2011, 29(7): 644-652.
doi: 10.1038/nbt.1883 pmid: 21572440 |
| [15] |
Altschul SF, Madden TL, Schäffer AA, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs[J]. Nucleic Acids Res, 1997, 25(17): 3389-3402.
doi: 10.1093/nar/25.17.3389 pmid: 9254694 |
| [16] |
Xie C, Mao XZ, Huang JJ, et al. KOBAS 2.0: a web server for annotation and identification of enriched pathways and diseases[J]. Nucleic Acids Res, 2011, 39(Web Server issue): W316-W322.
doi: 10.1093/nar/gkr483 URL |
| [17] |
Eddy SR. HMMER: profile HMMs for protein sequence analysis. Bioinformatics, 1998, 14: 755-763.
doi: 10.1093/bioinformatics/14.9.755 pmid: 9918945 |
| [18] |
Yu GC, Wang LG, Han YY, et al. clusterProfiler: an R package for comparing biological themes among gene clusters[J]. OMICS, 2012, 16(5): 284-287.
doi: 10.1089/omi.2011.0118 pmid: 22455463 |
| [19] |
Langmead B, Trapnell C, Pop M, et al. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome[J]. Genome Biol, 2009, 10(3): R25.
doi: 10.1186/gb-2009-10-3-r25 URL |
| [20] |
Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome[J]. BMC Bioinformatics, 2011, 12: 323.
doi: 10.1186/1471-2105-12-323 pmid: 21816040 |
| [41] |
Osnato M, Cota I, Nebhnani P, et al. Photoperiod control of plant growth: flowering time genes beyond flowering[J]. Front Plant Sci, 2022, 12: 805635.
doi: 10.3389/fpls.2021.805635 URL |
| [42] |
Andrés J, Caruana J, Liang JH, et al. Woodland strawberry axillary bud fate is dictated by a crosstalk of environmental and endogenous factors[J]. Plant Physiol, 2021, 187(3): 1221-1234.
doi: 10.1093/plphys/kiab421 pmid: 34618090 |
| [43] |
Zhou TT, Song BT, Liu TF, et al. Phytochrome F plays critical roles in potato photoperiodic tuberization[J]. Plant J, 2019, 98(1): 42-54.
doi: 10.1111/tpj.14198 |
| [21] |
Kumar L, Futschik ME. Mfuzz: a software package for soft clustering of microarray data[J]. Bioinformation, 2007, 2(1): 5-7.
doi: 10.6026/97320630002005 pmid: 18084642 |
| [22] |
Chen CJ, Chen H, Zhang Y, et al. TBtools: an integrative toolkit developed for interactive analyses of big biological data[J]. Mol Plant, 2020, 13(8): 1194-1202.
doi: S1674-2052(20)30187-8 pmid: 32585190 |
| [23] |
Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis[J]. BMC Bioinformatics, 2008, 9: 559.
doi: 10.1186/1471-2105-9-559 pmid: 19114008 |
| [24] | Hu RB, Yu CJ, Wang XY, et al. De novo transcriptome analysis of Miscanthus lutarioriparius identifies candidate genes in rhizome development[J]. Front Plant Sci, 2017, 8: 492. |
| [25] |
Huang H, Wang Y, Wang SL, et al. Transcriptome-wide survey and expression analysis of stress-responsive NAC genes in Chrysanthemum lavandulifolium[J]. Plant Sci, 2012, 193/194: 18-27.
doi: 10.1016/j.plantsci.2012.05.004 URL |
| [26] |
Chai Q, Jin F, Merewitz E, et al. Growth and physiological traits associated with drought survival and post-drought recovery in perennial turfgrass species[J]. J Amer Soc Hort Sci, 2010, 135(2): 125-133.
doi: 10.21273/JASHS.135.2.125 URL |
| [27] | 张黎黎. 菊花DEAD-box RNA解旋酶基因CmRH56调节地下茎发生与干旱耐性的功能分析[D]. 北京: 中国农业大学, 2017. |
| Zhang LL. Function analysis of DEAD-box RNA helicase CmRH56 regulating morphogenesis of suckers and drought stress tolerance in Chrysanthemum[D]. Beijing: China Agricultural University, 2017. | |
| [28] |
Zhou YB, Chen M, Guo JK, et al. Overexpression of soybean DREB1 enhances drought stress tolerance of transgenic wheat in the field[J]. J Exp Bot, 2020, 71(6): 1842-1857.
doi: 10.1093/jxb/erz569 URL |
| [29] |
Wang GD, Xu XP, Wang H, et al. A tomato transcription factor, SlDREB3 enhances the tolerance to chilling in transgenic tomato[J]. Plant Physiol Biochem, 2019, 142: 254-262.
doi: 10.1016/j.plaphy.2019.07.017 URL |
| [30] |
Lim J, Lim CW, Lee SC. The pepper late embryogenesis abundant protein, CaDIL1, positively regulates drought tolerance and ABA signaling[J]. Front Plant Sci, 2018, 9: 1301.
doi: 10.3389/fpls.2018.01301 pmid: 30233631 |
| [31] | 袁娅娟. 草地早熟禾根茎扩展与内源激素及碳氮代谢随生育时期的动态变化[D]. 兰州: 甘肃农业大学, 2021. |
| Yuan YJ. Study on the dynamic changes of rhizome expansion, endogenous hormones, carbon and nitrogen metabolism of Poa pratensis during growth stage[D]. Lanzhou: Gansu Agricultural University, 2021. | |
| [32] |
Li LS, Xia TZ, Li B, et al. Hormone and carbohydrate metabolism associated genes play important roles in rhizome bud full-year germination of Cephalostachyum pingbianense[J]. Physiol Plant, 2022, 174(2): e13674.
doi: 10.1111/ppl.v174.2 URL |
| [33] |
Ma XQ, Huang BR. Gibberellin-stimulation of rhizome elongation and differential GA-responsive proteomic changes in two grass species[J]. Front Plant Sci, 2016, 7: 905.
doi: 10.3389/fpls.2016.00905 pmid: 27446135 |
| [34] |
Zdunek-Zastocka E, Grabowska A. The interplay of PsABAUGT1 with other abscisic acid metabolic genes in the regulation of ABA homeostasis during the development of pea seeds and germination in the presence of H2O2[J]. Plant Sci, 2019, 285: 79-90.
doi: S0168-9452(19)30178-5 pmid: 31203896 |
| [35] |
Zheng CL, Acheampong AK, Shi ZW, et al. Abscisic acid catabolism enhances dormancy release of grapevine buds[J]. Plant Cell Environ, 2018, 41(10): 2490-2503.
doi: 10.1111/pce.v41.10 URL |
| [36] | Wang J, Xu YD, Yin ZN, et al. Overexpression of the persimmon abscisic acid DkUGT3 gene alters plant/fruit development in transgenic tomato[J]. J Plant Growth Regul, 2023: 1-15. |
| [37] |
Liang B, Zheng Y, Wang J, et al. Overexpression of the persimmon abscisic acid β-glucosidase gene(DkBG1)alters fruit ripening in transgenic tomato[J]. Plant J, 2020, 102(6): 1220-1233.
doi: 10.1111/tpj.v102.6 URL |
| [38] |
Masuda JI, Ozaki Y, Okubo H. Rhizome transition to storage organ is under phytochrome control in lotus(Nelumbo nucifera)[J]. Planta, 2007, 226(4): 909-915.
doi: 10.1007/s00425-007-0536-9 URL |
| [39] |
Saxena P, Huang BR, Bonos SA, et al. Photoperiod and temperature effects on rhizome production and tillering rate in tall fescue[Lolium arundinaceum(schreb.) darby.[J]. Crop Sci, 2014, 54(3): 1205-1210.
doi: 10.2135/cropsci2013.08.0565 URL |
| [40] |
Xu ZE, Chen HJ, Ji LF, et al. Polymorphisms of the FT gene as a tool to identify underground rhizome types of bamboos[J]. Euphytica, 2017, 213(1): 25.
doi: 10.1007/s10681-016-1824-x |
| [1] | LIN Hong-yan, GUO Xiao-rui, LIU Di, LI Hui, LU Hai. Molecular Mechanism of Transcriptional Factor AtbHLH68 in Regulating Cell Wall Development by Transcriptome Analysis [J]. Biotechnology Bulletin, 2023, 39(9): 105-116. |
| [2] | MIAO Yong-mei, MIAO Cui-ping, YU Qing-cai. Properties of Bacillus subtilis Strain BBs-27 Fermentation Broth and the Inhibition of Lipopeptides Against Fusarium culmorum [J]. Biotechnology Bulletin, 2023, 39(9): 255-267. |
| [3] | LIU Bao-cai, CHEN Jing-ying, ZHANG Wu-jun, HUANG Ying-zhen, ZHAO Yun-qing, LIU Jian-chao, WEI Zhi-cheng. Characteristics Analysis of Seed Microrhizome Gene Expression of Polygonatum cyrtonema [J]. Biotechnology Bulletin, 2023, 39(8): 220-233. |
| [4] | FU Yu, JIA Rui-rui, HE He, WANG Liang-gui, YANG Xiu-lian. Growth Differences Among Grafted Seedlings with Two Rootstocks of Catalpa bungei and Comparative Analysis of Transcriptome [J]. Biotechnology Bulletin, 2023, 39(8): 251-261. |
| [5] | KONG De-zhen, DUAN Zhen-yu, WANG Gang, ZHANG Xin, XI Lin-qiao. Physiological Characteristics and Transcriptome Analysis of Sorghum bicolor × S. Sudanense Seedlings Under Salt-alkali Stress [J]. Biotechnology Bulletin, 2023, 39(6): 199-207. |
| [6] | LIU Hui, LU Yang, YE Xi-miao, ZHOU Shuai, LI Jun, TANG Jian-bo, CHEN En-fa. Comparative Transcriptome Analysis of Cadmium Stress Response Induced by Exogenous Sulfur in Tartary Buckwheat [J]. Biotechnology Bulletin, 2023, 39(5): 177-191. |
| [7] | XIE Yang, XING Yu-meng, ZHOU Guo-yan, LIU Mei-yan, YIN Shan-shan, YAN Li-ying. Transcriptome Analysis of Diploid and Autotetraploid in Cucumber Fruit [J]. Biotechnology Bulletin, 2023, 39(3): 152-162. |
| [8] | HU Li-li, LIN Bo-rong, WANG Hong-hong, CHEN Jian-song, LIAO Jin-ling, ZHUO Kan. Transcriptome and Candidate Effectors Analysis of Pratylenchus brachyurus [J]. Biotechnology Bulletin, 2023, 39(3): 254-266. |
| [9] | SUN Yan-qiu, XIE Cai-yun, TANG Yue-qin. Construction and Mechanism Analysis of High-temperature Resistant Saccharomyces cerevisiae [J]. Biotechnology Bulletin, 2023, 39(11): 226-237. |
| [10] | LUO Hao-tian, WANG Long, WANG Yu-qian, WANG Yue, LI Jia-zhen, YANG Meng-ke, ZHANG Jie, DENG Xin, WANG Hong-yan. Genome-wide Identification and Expression Analysis of the RNAi-related Gene Families in Setaria viridis [J]. Biotechnology Bulletin, 2023, 39(1): 175-186. |
| [11] | XIN Jian-pan, LI Yan, ZHAO Chu, TIAN Ru-nan. Transcriptome Sequencing in the Leaves of Pontederia cordata with Cadmium Exposure and Gene Mining in Phenypropanoid Pathways [J]. Biotechnology Bulletin, 2022, 38(6): 198-210. |
| [12] | XU Jin, LI Tao, LI Chu-lin, ZHU Shun-ni, WANG Zhong-ming, XIANG Wen-zhou. Effects of Temperature on the Growth,Total Lipid and Eicosapentaenoic Acid Synthesis of Eustigmatos sp. [J]. Biotechnology Bulletin, 2022, 38(6): 261-271. |
| [13] | XIONG He-li, SHA Qian, LIU Shao-na, XIANG De-cai, ZHANG Bin, ZHAO Zhi-yong. Application of Single-cell Transcriptome Sequencing in Animals [J]. Biotechnology Bulletin, 2022, 38(3): 226-233. |
| [14] | ZHANG Bin, YANG Xin-xia. Identification of Key Transcription Factors in Response to Salt Stress in Rice [J]. Biotechnology Bulletin, 2022, 38(3): 9-15. |
| [15] | GUAN Yi, LI Xin, WANG Ding-yi, DU Xi, ZHANG Long-bin, YE Xiu-yun. Functional Study of BbRho5 on the Growth Rate of Beauveria bassiana [J]. Biotechnology Bulletin, 2022, 38(2): 132-140. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||