








Biotechnology Bulletin ›› 2023, Vol. 39 ›› Issue (2): 147-160.doi: 10.13560/j.cnki.biotech.bull.1985.2022-0746
Previous Articles Next Articles
LV Yu-jing1( ), WU Dan-dan1, KONG Chun-yan1,2, YANG Yu1, GONG Ming1(
), WU Dan-dan1, KONG Chun-yan1,2, YANG Yu1, GONG Ming1( )
)
Received:2022-06-21
Online:2023-02-26
Published:2023-03-07
LV Yu-jing, WU Dan-dan, KONG Chun-yan, YANG Yu, GONG Ming. Genome-wide Identification of XTH Gene Family and Their Interacting miRNAs and Possible Roles in Low Temperature Adaptation in Jatropha curcas L.[J]. Biotechnology Bulletin, 2023, 39(2): 147-160.
| 引物名称Primer name | 引物序列Primer sequence(5'-3') |
|---|---|
| JcXTH2-F | GAGAGCAACAATTCCACCTTTG |
| JcXTH2-R | GGGTAAGGAACACCGTTTGA |
| JcXTH3-F | CTCCCTTCACTGCTTCTTACAG |
| JcXTH3-R | CACCCATTTGAGCCTCTCTT |
| JcXTH18-F | CTACCAGGGAATACCACACTTAC |
| JcXTH18-R | CGTTCCACAGGCTTGAGTATAA |
| JcXTH19-F | GTGCACAAGAACAATGGAGAAC |
| JcXTH19-R | TGACAACCATCGACGCTAAA |
| JcXTH23-F | GCAACAAGAGGTGGACTAGAA |
| JcXTH23-R | CATGGCACTTAGGGCTTGATA |
| JcXTH27-F | TACTGTTGGGCGCGATAATG |
| JcXTH27-R | CCAGACCCAGAAACCCTATCTA |
| GAPDH-F | TGAAGGACTGGAGAGGTGGAAGAGC |
| GAPDH-R | ATCAACAGTTGGAACACGGAAAGCC |
Table 1 Related primers for RT-qPCR analysis
| 引物名称Primer name | 引物序列Primer sequence(5'-3') |
|---|---|
| JcXTH2-F | GAGAGCAACAATTCCACCTTTG |
| JcXTH2-R | GGGTAAGGAACACCGTTTGA |
| JcXTH3-F | CTCCCTTCACTGCTTCTTACAG |
| JcXTH3-R | CACCCATTTGAGCCTCTCTT |
| JcXTH18-F | CTACCAGGGAATACCACACTTAC |
| JcXTH18-R | CGTTCCACAGGCTTGAGTATAA |
| JcXTH19-F | GTGCACAAGAACAATGGAGAAC |
| JcXTH19-R | TGACAACCATCGACGCTAAA |
| JcXTH23-F | GCAACAAGAGGTGGACTAGAA |
| JcXTH23-R | CATGGCACTTAGGGCTTGATA |
| JcXTH27-F | TACTGTTGGGCGCGATAATG |
| JcXTH27-R | CCAGACCCAGAAACCCTATCTA |
| GAPDH-F | TGAAGGACTGGAGAGGTGGAAGAGC |
| GAPDH-R | ATCAACAGTTGGAACACGGAAAGCC |
| 基因名称Gene name | 基因ID Gene ID | 氨基酸数目 Number of amino acids/aa | 等电点Isoelectric point | 分子量 Molecular weight/kD | 亲水性平均值 GRAVY | 亚细胞定位 Subcellular localization |
|---|---|---|---|---|---|---|
| JcXTH1 | XM_012218696.1 | 285 | 6.42 | 32.36735 | -0.394 | 细胞壁, 细胞质 |
| JcXTH2 | XM_012218694.1 | 269 | 9.04 | 30.94703 | -0.438 | 细胞壁, 细胞质 |
| JcXTH3 | XM_012218698.1 | 286 | 6.14 | 32.08499 | -0.319 | 细胞壁, 细胞质 |
| JcXTH4 | XM_012218697.1 | 286 | 5.47 | 32.20504 | -0.353 | 细胞壁, 细胞质 |
| JcXTH5 | XM_012218699.1 | 283 | 5.21 | 31.86363 | -0.405 | 细胞壁, 细胞质 |
| JcXTH6 | XM_012218701.1 | 283 | 5.1 | 31.8406 | -0.353 | 细胞壁, 细胞质 |
| JcXTH7 | XM_012218700.1 | 283 | 5.18 | 31.72839 | -0.367 | 细胞壁, 细胞质 |
| JcXTH8 | XM_012210833.1 | 283 | 5.79 | 31.90475 | -0.296 | 细胞壁, 细胞质 |
| JcXTH9 | XM_012218702.1 | 299 | 5.43 | 34.08454 | -0.219 | 细胞壁, 细胞质 |
| JcXTH10 | XM_012210835.1 | 279 | 7.61 | 31.34916 | -0.36 | 细胞壁, 细胞质 |
| JcXTH11 | XM_012222502.1 | 288 | 8.78 | 32.76008 | -0.301 | 细胞壁, 细胞质 |
| JcXTH12 | XM_012227056.1 | 294 | 9.05 | 33.9776 | -0.386 | 细胞壁 |
| JcXTH13 | XM_012209320.1 | 293 | 6.64 | 32.91467 | -0.514 | 细胞壁 |
| JcXTH14 | XM_012216981.1 | 323 | 6.76 | 36.95264 | -0.393 | 细胞壁 |
| JcXTH15 | XM_012225387.1 | 341 | 8.73 | 39.63884 | -0.51 | 细胞壁 |
| JcXTH16 | XM_012229959.1 | 311 | 6.41 | 34.99533 | -0.326 | 细胞壁 |
| JcXTH17 | XM_012233283.1 | 293 | 8.45 | 34.01056 | -0.452 | 细胞壁, 细胞质 |
| JcXTH18 | XM_012212020.1 | 293 | 6.99 | 34.10647 | -0.439 | 细胞壁, 细胞质 |
| JcXTH19 | XM_012236012.1 | 299 | 5 | 34.8449 | -0.6 | 细胞壁 |
| JcXTH20 | XM_012218715.1 | 269 | 6.31 | 30.87093 | -0.361 | 细胞壁 |
| JcXTH21 | XM_012230444.1 | 284 | 6.44 | 32.68983 | -0.361 | 细胞壁 |
| JcXTH22 | XM_012215879.1 | 293 | 4.88 | 32.97573 | -0.353 | 细胞壁 |
| JcXTH23 | XM_012224087.1 | 292 | 6.7 | 33.62413 | -0.298 | 细胞壁 |
| JcXTH24 | XM_012223597.1 | 288 | 5.37 | 32.77481 | -0.297 | 细胞壁 |
| JcXTH25 | XM_012217377.1 | 294 | 8.89 | 34.70349 | -0.382 | 细胞壁 |
| JcXTH26 | XM_012221356.1 | 291 | 8.51 | 33.28685 | -0.225 | 细胞壁 |
| JcXTH27 | XM_012223052.1 | 277 | 8.82 | 31.58769 | -0.288 | 细胞壁, 细胞质 |
| JcXTH28 | XM_012219356.1 | 291 | 8.57 | 33.20157 | -0.282 | 细胞壁, 细胞质 |
| JcXTH29 | XM_012237054.1 | 288 | 9.03 | 32.9001 | -0.427 | 细胞壁 |
Table 2 Physicochemical properties and subcellular location of identified XTH proteins in J. curcas
| 基因名称Gene name | 基因ID Gene ID | 氨基酸数目 Number of amino acids/aa | 等电点Isoelectric point | 分子量 Molecular weight/kD | 亲水性平均值 GRAVY | 亚细胞定位 Subcellular localization |
|---|---|---|---|---|---|---|
| JcXTH1 | XM_012218696.1 | 285 | 6.42 | 32.36735 | -0.394 | 细胞壁, 细胞质 |
| JcXTH2 | XM_012218694.1 | 269 | 9.04 | 30.94703 | -0.438 | 细胞壁, 细胞质 |
| JcXTH3 | XM_012218698.1 | 286 | 6.14 | 32.08499 | -0.319 | 细胞壁, 细胞质 |
| JcXTH4 | XM_012218697.1 | 286 | 5.47 | 32.20504 | -0.353 | 细胞壁, 细胞质 |
| JcXTH5 | XM_012218699.1 | 283 | 5.21 | 31.86363 | -0.405 | 细胞壁, 细胞质 |
| JcXTH6 | XM_012218701.1 | 283 | 5.1 | 31.8406 | -0.353 | 细胞壁, 细胞质 |
| JcXTH7 | XM_012218700.1 | 283 | 5.18 | 31.72839 | -0.367 | 细胞壁, 细胞质 |
| JcXTH8 | XM_012210833.1 | 283 | 5.79 | 31.90475 | -0.296 | 细胞壁, 细胞质 |
| JcXTH9 | XM_012218702.1 | 299 | 5.43 | 34.08454 | -0.219 | 细胞壁, 细胞质 |
| JcXTH10 | XM_012210835.1 | 279 | 7.61 | 31.34916 | -0.36 | 细胞壁, 细胞质 |
| JcXTH11 | XM_012222502.1 | 288 | 8.78 | 32.76008 | -0.301 | 细胞壁, 细胞质 |
| JcXTH12 | XM_012227056.1 | 294 | 9.05 | 33.9776 | -0.386 | 细胞壁 |
| JcXTH13 | XM_012209320.1 | 293 | 6.64 | 32.91467 | -0.514 | 细胞壁 |
| JcXTH14 | XM_012216981.1 | 323 | 6.76 | 36.95264 | -0.393 | 细胞壁 |
| JcXTH15 | XM_012225387.1 | 341 | 8.73 | 39.63884 | -0.51 | 细胞壁 |
| JcXTH16 | XM_012229959.1 | 311 | 6.41 | 34.99533 | -0.326 | 细胞壁 |
| JcXTH17 | XM_012233283.1 | 293 | 8.45 | 34.01056 | -0.452 | 细胞壁, 细胞质 |
| JcXTH18 | XM_012212020.1 | 293 | 6.99 | 34.10647 | -0.439 | 细胞壁, 细胞质 |
| JcXTH19 | XM_012236012.1 | 299 | 5 | 34.8449 | -0.6 | 细胞壁 |
| JcXTH20 | XM_012218715.1 | 269 | 6.31 | 30.87093 | -0.361 | 细胞壁 |
| JcXTH21 | XM_012230444.1 | 284 | 6.44 | 32.68983 | -0.361 | 细胞壁 |
| JcXTH22 | XM_012215879.1 | 293 | 4.88 | 32.97573 | -0.353 | 细胞壁 |
| JcXTH23 | XM_012224087.1 | 292 | 6.7 | 33.62413 | -0.298 | 细胞壁 |
| JcXTH24 | XM_012223597.1 | 288 | 5.37 | 32.77481 | -0.297 | 细胞壁 |
| JcXTH25 | XM_012217377.1 | 294 | 8.89 | 34.70349 | -0.382 | 细胞壁 |
| JcXTH26 | XM_012221356.1 | 291 | 8.51 | 33.28685 | -0.225 | 细胞壁 |
| JcXTH27 | XM_012223052.1 | 277 | 8.82 | 31.58769 | -0.288 | 细胞壁, 细胞质 |
| JcXTH28 | XM_012219356.1 | 291 | 8.57 | 33.20157 | -0.282 | 细胞壁, 细胞质 |
| JcXTH29 | XM_012237054.1 | 288 | 9.03 | 32.9001 | -0.427 | 细胞壁 |
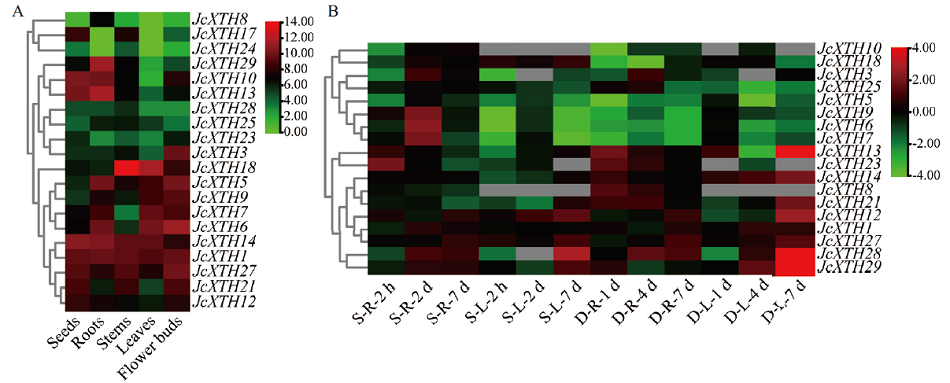
Fig. 7 Expression profiles of JcXTH genes in different organs(A)and under salt and drought stress(B) S: Salt stress; D: drought stress; R: root; L: leaf

Fig. 8 RT-qPCR analysis of the genes responding to chill-hardening in JcXTH gene family of J. curcas during chill-hardening at 12℃ Different lowercase letters on the histogram indicate significant differences(P<0.05)
| [1] |
Cavalier DM, Lerouxel O, Neumetzler L, et al. Disrupting two Arabidopsis thaliana xylosyltransferase genes results in plants deficient in xyloglucan, a major primary cell wall component[J]. Plant Cell, 2008, 20(6): 1519-1537.
doi: 10.1105/tpc.108.059873 pmid: 18544630 |
| [2] |
Pauly M, Gille S, Liu LF, et al. Hemicellulose biosynthesis[J]. Planta, 2013, 238(4): 627-642.
doi: 10.1007/s00425-013-1921-1 pmid: 23801299 |
| [3] | Schultink A, Liu LF, Zhu L, et al. Structural diversity and function of xyloglucan sidechain substituents[J]. Plants(Basel), 2014, 3(4): 526-542. |
| [4] |
Hayashi T, Kaida RM. Functions of xyloglucan in plant cells[J]. Mol Plant, 2011, 4(1): 17-24.
doi: 10.1093/mp/ssq063 pmid: 20943810 |
| [5] |
Jiang Y, Li YH, Lu C, et al. Isolation and characterization of Populus xyloglucan endotransglycosylase/hydrolase(XTH)involved in osmotic stress responses[J]. Int J Biol Macromol, 2020, 155: 1277-1287.
doi: S0141-8130(19)35369-3 pmid: 31730960 |
| [6] |
Stratilová B, Kozmon S, Stratilová E, et al. Plant xyloglucan xyloglucosyl transferases and the cell wall structure: subtle but significant[J]. Molecules, 2020, 25(23): 5619.
doi: 10.3390/molecules25235619 URL |
| [7] | Cosgrove DJ. Catalysts of plant cell wall loosening[J]. F1000Res, 2016, 5: F1000 Faculty Rev-F1000 Faculty 119. |
| [8] |
McGregor N, Yin V, Tung CC, et al. Crystallographic insight into the evolutionary origins of xyloglucan endotransglycosylases and endohydrolases[J]. Plant J, 2017, 89(4): 651-670.
doi: 10.1111/tpj.13421 URL |
| [9] |
Yokoyama R, Nishitani K. A comprehensive expression analysis of all members of a gene family encoding cell-wall enzymes allowed us to predict Cis-regulatory regions involved in cell-wall construction in specific organs of Arabidopsis[J]. Plant Cell Physiol, 2001, 42(10): 1025-1033.
pmid: 11673616 |
| [10] |
Yokoyama R, Rose JKC, Nishitani K. A surprising diversity and abundance of xyloglucan endotransglucosylase/hydrolases in rice. Classification and expression analysis[J]. Plant Physiol, 2004, 134(3): 1088-1099.
pmid: 14988479 |
| [11] |
Wang M, Xu ZC, Ding AM, et al. Genome-wide identification and expression profiling analysis of the xyloglucan endotransglucosylase/hydrolase gene family in tobacco(Nicotiana tabacum L.)[J]. Genes, 2018, 9(6): 273.
doi: 10.3390/genes9060273 URL |
| [12] | Song L, Valliyodan B, Prince S, et al. Characterization of the XTH gene family: new insight to the roles in soybean flooding tolerance[J]. Int J Mol Sci, 2018, 19(9): E2705. |
| [13] |
Fu MM, Liu C, Wu FB. Genome-wide identification, characterization and expression analysis of xyloglucan endotransglucosylase/hydrolase genes family in barley(Hordeum vulgare)[J]. Molecules, 2019, 24(10): 1935.
doi: 10.3390/molecules24101935 URL |
| [14] |
Kaewthai N, Gendre D, Eklöf JM, et al. Group III-A XTH genes of Arabidopsis encode predominant xyloglucan endohydrolases that are dispensable for normal growth[J]. Plant Physiol, 2013, 161(1): 440-454.
doi: 10.1104/pp.112.207308 pmid: 23104861 |
| [15] |
Zhu XF, Shi YZ, Lei GJ, et al. XTH31, encoding an in vitro XEH/XET-active enzyme, regulates aluminum sensitivity by modulating in vivo XET action, cell wall xyloglucan content, and aluminum binding capacity in Arabidopsis[J]. Plant Cell, 2012, 24(11): 4731-4747.
doi: 10.1105/tpc.112.106039 URL |
| [16] |
Harada T, Torii Y, Morita S, et al. Cloning, characterization, and expression of xyloglucan endotransglucosylase/hydrolase and expansin genes associated with petal growth and development during carnation flower opening[J]. J Exp Bot, 2011, 62(2): 815-823.
doi: 10.1093/jxb/erq319 pmid: 20959626 |
| [17] |
Liu YB, Lu SM, Zhang JF, et al. A xyloglucan endotransglucosylase/hydrolase involves in growth of primary root and alters the deposition of cellulose in Arabidopsis[J]. Planta, 2007, 226(6): 1547-1560.
doi: 10.1007/s00425-007-0591-2 URL |
| [18] |
Jiménez T, Martín I, Labrador E, et al. The immunolocation of a xyloglucan endotransglucosylase/hydrolase specific to elongating tissues in Cicer arietinum suggests a role in the elongation of vascular cells[J]. J Exp Bot, 2006, 57(15): 3979-3988.
pmid: 17075081 |
| [19] | le Gall H, Philippe F, Domon JM, et al. Cell wall metabolism in response to abiotic stress[J]. Plants(Basel), 2015, 4(1): 112-166. |
| [20] |
Cho SK, Kim JE, Park JA, et al. Constitutive expression of abiotic stress-inducible hot pepper CaXTH3, which encodes a xyloglucan endotransglucosylase/hydrolase homolog, improves drought and salt tolerance in transgenic Arabidopsis plants[J]. FEBS Lett, 2006, 580(13): 3136-3144.
doi: 10.1016/j.febslet.2006.04.062 URL |
| [21] |
Shi HT, Ye TT, Zhong B, et al. AtHAP5A modulates freezing stress resistance in Arabidopsis through binding to CCAAT motif of AtXTH21[J]. New Phytol, 2014, 203(2): 554-567.
doi: 10.1111/nph.12812 URL |
| [22] |
Han YS, Sa G, Sun J, et al. Overexpression of Populus euphratica xyloglucan endotransglucosylase/hydrolase gene confers enhanced cadmium tolerance by the restriction of root cadmium uptake in transgenic tobacco[J]. Environ Exp Bot, 2014, 100: 74-83.
doi: 10.1016/j.envexpbot.2013.12.021 URL |
| [23] |
Han YS, Wang W, Sun J, et al. Populus euphratica XTH overexpression enhances salinity tolerance by the development of leaf succulence in transgenic tobacco plants[J]. J Exp Bot, 2013, 64(14): 4225-4238.
doi: 10.1093/jxb/ert229 URL |
| [24] |
Lama AD, Klemola T, Saloniemi I, et al. Factors affecting genetic and seed yield variability of Jatropha curcas(L.)across the globe: a review[J]. Energy Sustain Dev, 2018, 42: 170-182.
doi: 10.1016/j.esd.2017.09.002 URL |
| [25] |
Maghuly F, Laimer M. Jatropha curcas, a biofuel crop: functional genomics for understanding metabolic pathways and genetic improvement[J]. Biotechnol J, 2013, 8(10): 1172-1182.
doi: 10.1002/biot.201300231 pmid: 24092674 |
| [26] |
Pompelli MF, Ferreira PPB, Chaves ARM, et al. Physiological, metabolic, and stomatal adjustments in response to salt stress in Jatropha curcas[J]. Plant Physiol Biochem, 2021, 168: 116-127.
doi: 10.1016/j.plaphy.2021.09.039 URL |
| [27] |
Yang SL, Lan SS, Deng FF, et al. Effects of calcium and calmodulin antagonists on chilling stress-induced proline accumulation in Jatropha curcas L[J]. J Plant Growth Regul, 2016, 35(3): 815-826.
doi: 10.1007/s00344-016-9584-3 URL |
| [28] |
Ao PX, Li ZG, Fan DM, et al. Involvement of antioxidant defense system in chill hardening-induced chilling tolerance in Jatropha curcas seedlings[J]. Acta Physiol Plant, 2013, 35(1): 153-160.
doi: 10.1007/s11738-012-1058-z URL |
| [29] |
Ao PX, Li ZG, Gong M. Involvement of compatible solutes in chill hardening-induced chilling tolerance in Jatropha curcas seedlings[J]. Acta Physiol Plant, 2013, 35(12): 3457-3464.
doi: 10.1007/s11738-013-1381-z URL |
| [30] |
张翠桔, 莫蓓莘, 陈雪梅, 等. 植物miRNA作用方式的分子机制研究进展[J]. 生物技术通报, 2020, 36(7): 1-14.
doi: 10.13560/j.cnki.biotech.bull.1985.2020-0262 |
| Zhang CJ, Mo BX, Chen XM, et al. Advances on the molecular action mechanisms of plant miRNA[J]. Biotechnol Bull, 2020, 36(7): 1-14. | |
| [31] | 郝大海, 龚明. miRNA作用机制研究进展[J]. 基因组学与应用生物学, 2020, 39(8): 3647-3657. |
| Hao DH, Gong M. The progress of miRNA action mechanism[J]. Genom Appl Biol, 2020, 39(8): 3647-3657. | |
| [32] | Wang SS, Wang HB, Gong M. Identification of micrornas involved in chilling response by deep sequencing of Jatropha curcas L. small rnas at the global genome level[C]. Copenhagen: 21st European Biomass Conference and Exhibition, 2013. |
| [33] |
Wang HB, Zou ZR, Wang SS, et al. Global analysis of transcriptome responses and gene expression profiles to cold stress of Jatropha curcas L[J]. PLoS One, 2013, 8(12): e82817.
doi: 10.1371/journal.pone.0082817 URL |
| [34] |
吴丹丹, 陈永坤, 杨宇, 等. 小桐子半胱氨酸蛋白酶家族和相应miRNAs的鉴定及其对低温锻炼的响应[J]. 植物学报, 2021, 56(5): 544-558.
doi: 10.11983/CBB21014 |
| Wu DD, Chen YK, Yang Y, et al. Identification of the cysteine protease family and corresponding miRNAs in Jatropha curcas and their response to chill-hardening[J]. Chin Bull Bot, 2021, 56(5): 544-558. | |
| [35] | 吴丹丹, 陈永坤, 杨宇, 等. 小桐子cystatin家族基因和相应miRNAs的鉴定及其在低温响应中可能的作用[J]. 植物生理学报, 2021, 57(2): 347-361. |
|
Wu DD, Chen YK, Yang Y, et al. Identification of cystatin family genes and their corresponding miRNAs in Jatropha curcas and possible roles in response to low temperature[J]. Plant Physiol J, 2021, 57(2): 347-361.
doi: 10.1104/pp.57.3.347 URL |
|
| [36] | 李忠光, 龚明. 不同化学消毒剂对小桐子种子萌发和幼苗生长的影响[J]. 种子, 2011, 30(2): 4-7, 12. |
| Li ZG, Gong M. Effects of different chemical disinfectant on seed germination and seedling growth of Jatropha curcas L[J]. Seed, 2011, 30(2): 4-7, 12. | |
| [37] |
Zhang X, Pan BZ, Chen MS, et al. JCDB: a comprehensive knowledge base for Jatropha curcas, an emerging model for woody energy plants[J]. BMC Genomics, 2019, 20(Suppl 9): 958.
doi: 10.1186/s12864-019-6356-z pmid: 31874631 |
| [38] |
Marchler-Bauer A, Bo Y, Han LY, et al. CDD/SPARCLE: functional classification of proteins via subfamily domain architectures[J]. Nucleic Acids Res, 2017, 45(D1): D200-D203.
doi: 10.1093/nar/gkw1129 URL |
| [39] |
Bailey TL, Boden M, Buske FA, et al. MEME SUITE: tools for motif discovery and searching[J]. Nucleic Acids Res, 2009, 37(Web Server issue): W202-W208.
doi: 10.1093/nar/gkp335 URL |
| [40] |
Wu PZ, Zhou CP, Cheng SF, et al. Integrated genome sequence and linkage map of physic nut(Jatropha curcasL.), a biodiesel plant[J]. Plant J, 2015, 81(5): 810-821.
doi: 10.1111/tpj.12761 URL |
| [41] |
孔春艳, 陈永坤, 王莎莎, 等. 小桐子低温胁迫下microRNA实时荧光定量PCR内参的筛选与比较[J]. 生物技术通报, 2019, 35(7): 25-31.
doi: 10.13560/j.cnki.biotech.bull.1985.2019-0042 |
| Kong CY, Chen YK, Wang SS, et al. Screening and comparison of reference genes for microRNA quantitative real-time PCR in Jatropha curcas under chilling stress[J]. Biotechnol Bull, 2019, 35(7): 25-31. | |
| [42] |
Opazo MC, Lizana R, Stappung Y, et al. XTHs from Fragaria vesca: genomic structure and transcriptomic analysis in ripening fruit and other tissues[J]. BMC Genomics, 2017, 18(1): 852.
doi: 10.1186/s12864-017-4255-8 URL |
| [43] |
Chebli Y, Geitmann A. Cellular growth in plants requires regulation of cell wall biochemistry[J]. Curr Opin Cell Biol, 2017, 44: 28-35.
doi: S0955-0674(17)30004-2 pmid: 28131101 |
| [44] |
Han Y, Han SK, Ban QY, et al. Overexpression of persimmon DkXTH1 enhanced tolerance to abiotic stress and delayed fruit softening in transgenic plants[J]. Plant Cell Rep, 2017, 36(4): 583-596.
doi: 10.1007/s00299-017-2105-4 pmid: 28155115 |
| [45] | Dong JL. Isolation of a novel xyloglucan endotransglucosylase(OsXET9)gene from rice and analysis of the response of this gene to abiotic stresses[J]. Afr J Biotechnol, 2011, 10(76): 17424-17434. |
| [46] |
Sasidharan R, Chinnappa CC, Staal M, et al. Light quality-mediated petiole elongation in Arabidopsis during shade avoidance involves cell wall modification by xyloglucan endotransglucosylase/hydrolases[J]. Plant Physiol, 2010, 154(2): 978-990.
doi: 10.1104/pp.110.162057 pmid: 20688978 |
| [47] |
Du HM, Hu XQ, Yang W, et al. ZmXTH, a xyloglucan endotransglucosylase/hydrolase gene of maize, conferred aluminum tolerance in Arabidopsis[J]. J Plant Physiol, 2021, 266: 153520.
doi: 10.1016/j.jplph.2021.153520 URL |
| [48] |
Xu PP, Fang S, Chen HY, et al. The brassinosteroid-responsive xyloglucan endotransglucosylase/hydrolase 19(XTH19)and XTH23 genes are involved in lateral root development under salt stress in Arabidopsis[J]. Plant J, 2020, 104(1): 59-75.
doi: 10.1111/tpj.14905 URL |
| [49] |
Pagano L, Rossi R, Paesano L, et al. miRNA regulation and stress adaptation in plants[J]. Environ Exp Bot, 2021, 184: 104369.
doi: 10.1016/j.envexpbot.2020.104369 URL |
| [1] | WEN Xiao-lei, LI Jian-yuan, LI Na, ZHANG Na, YANG Wen-xiang. Construction and Utilization of Yeast Two-hybrid cDNA Library of Wheat Interacted by Puccinia triticina [J]. Biotechnology Bulletin, 2023, 39(9): 136-146. |
| [2] | YANG Yang, ZHU Jin-cheng, LOU Hui, HAN Ze-gang, ZHANG Wei. Transcriptome Analysis of Interaction Between Gossypium barbadense and Fusarium oxysporum f. sp. vasinfectum [J]. Biotechnology Bulletin, 2023, 39(6): 259-273. |
| [3] | XIONG Shu-qi. Towards the Understanding on the Physiological Functions of Bile Acids and Interactions with Gut Microbiota [J]. Biotechnology Bulletin, 2023, 39(4): 187-200. |
| [4] | LI Kai-hang, WANG Hao-chen, CHENG Ke-xin, YANG Yan, JIN Yi, HE Xiao-qing. Genetic Mechanisms of Plant-microbiome Interaction by Genome-wide Association Analysis Study [J]. Biotechnology Bulletin, 2023, 39(2): 24-34. |
| [5] | LUO Ning, JIAO Yang, MAO Zhen-chuan, LI Hui-xia, XIE Bing-yan. Advances of Trichoderma in Controlling Root Knot Nematodes and Cyst Nematodes [J]. Biotechnology Bulletin, 2023, 39(2): 35-50. |
| [6] | YIN Guo-ying, LIU Chang, CHANG Yong-chun, YU Wang-jie, WANG Bing, ZHANG Pan, GUO Yu-shuang. Identification of the Cysteine Protease Family and Corresponding miRNAs in Nicotiana tabacum L. and Their Responses to PVY [J]. Biotechnology Bulletin, 2023, 39(10): 184-196. |
| [7] | SHEN Yue, TAO Bao-jie, HUA Xia, LV Bing, LIU Li-jun, CHEN Yun. Research Progress in the Interactions of Strigolactone with Hormones on Regulating Root Growth [J]. Biotechnology Bulletin, 2022, 38(8): 24-31. |
| [8] | ZHANG Bin, YANG Xin-xia. Identification of Key Transcription Factors in Response to Salt Stress in Rice [J]. Biotechnology Bulletin, 2022, 38(3): 9-15. |
| [9] | ZHANG Jun-feng, LI Meng-ke, WU Zhi-hao, CUI Xiao-long, XIAO wei, ZHANG Shi-ying. Effects of Bacteriophages DCEAV-31 and DCEIV-9 on the Algicidal Characteristics of Algicidal Bacterium Against Microcystis [J]. Biotechnology Bulletin, 2022, 38(11): 250-257. |
| [10] | TANG Xiao-li, JIANG Fu-dong, ZHANG Hong-xia. Research Progress in the Functions of SINA E3 Ubiquitin Ligase in Plant [J]. Biotechnology Bulletin, 2022, 38(10): 10-17. |
| [11] | CHEN Chen, HUANG Zhi-yang, YU Hai-yan, YUAN Hai-bin, TIAN Huai-xiang. Research Technology and Progress in Transcriptional Regulation in Prokaryotes [J]. Biotechnology Bulletin, 2022, 38(10): 54-65. |
| [12] | JIANG Yu-qi, SHU Xin-yue, ZHENG Ai-ping, WANG Ai-jun. Recent Progress in Molecular Mechanism of Interaction Between Rice and Tilletia horrida [J]. Biotechnology Bulletin, 2021, 37(9): 248-254. |
| [13] | LIU Juan, ZHU Chun-xiao, XIAO Xue-qiong, MO Chen-mi, WANG Gao-feng, XIAO Yan-nong. Screening of Protein Interacting with Purpureocillium lilacinum Cyclophilin PlCYP6 [J]. Biotechnology Bulletin, 2021, 37(7): 137-145. |
| [14] | HAN Shao-jie, ZHENG Jing-wu. Research Advances on the Functional Study of Host Resistance Genes to Heterodera glycines [J]. Biotechnology Bulletin, 2021, 37(7): 14-24. |
| [15] | LUO Wei, MU Qiong, SHU Jian-hong, WU Jia-hai, WANG Xiao-li. Expression,Protein Interactions and Biological Function Analysis of FaFT in Festuca arundinacea [J]. Biotechnology Bulletin, 2021, 37(4): 8-17. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||