








Biotechnology Bulletin ›› 2023, Vol. 39 ›› Issue (2): 254-262.doi: 10.13560/j.cnki.biotech.bull.1985.2022-0421
Previous Articles Next Articles
REN Si-yu1( ), JIANG Cong-yi1, YU Tao2, KANG Rui1, JIANG Xiao-bing1(
), JIANG Cong-yi1, YU Tao2, KANG Rui1, JIANG Xiao-bing1( )
)
Received:2022-04-07
Online:2023-02-26
Published:2023-03-07
REN Si-yu, JIANG Cong-yi, YU Tao, KANG Rui, JIANG Xiao-bing. Role of agr System in the Antimicrobial Resistance and Biofilm Formation of Listeria monocytogenes[J]. Biotechnology Bulletin, 2023, 39(2): 254-262.
| 引物名称 Primer name | 序列 Sequence(5'-3') | 长度 Length/bp |
|---|---|---|
| agrA-P1 | NNNNNNGGATCCGCTCTTACTGTCTTA- GCGTTCT | 882 |
| agrA-P2 | CCTGTGGCACCGATAAAATTCGCTGC- ATTCTGTTATCTTC | |
| agrA-P3 | GAAGATAACAGAATGCAGCGAATTTT- ATCGGTGCCACAGG | 568 |
| agrA-P4 | NNNNNNACGCGTAGTATTCCCATCGG- CATTT | |
| agrB/D-P1 | NNNNNNGGATCCGTCTACAAAGTTGA- TGGGATT | 680 |
| agrB/D-P2 | ACTCATAGAAGAATCCGCAAATCATCT- TTCCAGCGGTC | |
| agrB/D-P3 | GACCGCTGGAAAGATGATTTGCGGATT- CTTCTATGAGT | 608 |
| agrB/D-P4 | NNNNNNACGCGTGCTAAGACAGTAAG- AGCAACG |
Table 1 Primer sequences for PCR amplification
| 引物名称 Primer name | 序列 Sequence(5'-3') | 长度 Length/bp |
|---|---|---|
| agrA-P1 | NNNNNNGGATCCGCTCTTACTGTCTTA- GCGTTCT | 882 |
| agrA-P2 | CCTGTGGCACCGATAAAATTCGCTGC- ATTCTGTTATCTTC | |
| agrA-P3 | GAAGATAACAGAATGCAGCGAATTTT- ATCGGTGCCACAGG | 568 |
| agrA-P4 | NNNNNNACGCGTAGTATTCCCATCGG- CATTT | |
| agrB/D-P1 | NNNNNNGGATCCGTCTACAAAGTTGA- TGGGATT | 680 |
| agrB/D-P2 | ACTCATAGAAGAATCCGCAAATCATCT- TTCCAGCGGTC | |
| agrB/D-P3 | GACCGCTGGAAAGATGATTTGCGGATT- CTTCTATGAGT | 608 |
| agrB/D-P4 | NNNNNNACGCGTGCTAAGACAGTAAG- AGCAACG |

Fig. 2 Construction of ?agrA and ?agrB/D A: Amplification of upstream and downstream homology arms(M: DL2000 maker; 1: agrA upstream homology arm; 2: agrA downstream homology arm; 3: agrA fusion fragment; 4: agrB/D upstream homology arm; 5: agrB/D downstream homology arm; 6: agrB/D fusion fragment). B: Double digestion verification(M: DL2000; 1, 2: pMAD-agrA double digestion verification; 3, 4: pMAD-agrB/D double digestion verification). C: Knockout validation(M: DL2000; 1, 2: ΔagrA; 3: ΔagrB/D)
| 抗菌剂Antibacterial agents | EGD-e | ΔagrA | ΔagrB/D | ΔagrC | |
|---|---|---|---|---|---|
| 抗生素 Antibiotic | 氨苄西林 Ampicillin | 1 | 1 | 1 | 1 |
| 头孢噻肟 Cefotaxime | 12 | 8 | 12 | 12 | |
| 头孢噻吩 Cephalothin | 12 | 12 | 12 | 12 | |
| 红霉素 Erythromycin | 0.125 | 0.125 | 0.125 | 0.125 | |
| 卡那霉素 Kanamycin | 4 | 4 | 4 | 4 | |
| 四环素 Tetracycline | 0.5 | 0.5 | 0.5 | 0.5 | |
| 氯霉素 Chloramphenicol | 4 | 4 | 4 | 4 | |
| 环丙沙星 Ciprofloxacin | 1 | 1 | 0.5 | 0.5 | |
| 抗菌肽 Antimicrobial peptides | 乳酸链球菌素 Nisin | 10 | 10 | 10 | 10 |
| 消毒剂 Disinfectant | BC Benzalkonium chloride | 6 | 6 | 6 | 6 |
Table 2 MICs of antimicrobial agents against L. monocytogenes
| 抗菌剂Antibacterial agents | EGD-e | ΔagrA | ΔagrB/D | ΔagrC | |
|---|---|---|---|---|---|
| 抗生素 Antibiotic | 氨苄西林 Ampicillin | 1 | 1 | 1 | 1 |
| 头孢噻肟 Cefotaxime | 12 | 8 | 12 | 12 | |
| 头孢噻吩 Cephalothin | 12 | 12 | 12 | 12 | |
| 红霉素 Erythromycin | 0.125 | 0.125 | 0.125 | 0.125 | |
| 卡那霉素 Kanamycin | 4 | 4 | 4 | 4 | |
| 四环素 Tetracycline | 0.5 | 0.5 | 0.5 | 0.5 | |
| 氯霉素 Chloramphenicol | 4 | 4 | 4 | 4 | |
| 环丙沙星 Ciprofloxacin | 1 | 1 | 0.5 | 0.5 | |
| 抗菌肽 Antimicrobial peptides | 乳酸链球菌素 Nisin | 10 | 10 | 10 | 10 |
| 消毒剂 Disinfectant | BC Benzalkonium chloride | 6 | 6 | 6 | 6 |
| 菌株 Strain | 迟缓期LPD Lag phase duration/h | 平均最大生长速率MMGR Mean maximum growth rate/(Units·h-1) | 平均最大光密度值MMOD Mean maximum optical density/Units | ||||||
|---|---|---|---|---|---|---|---|---|---|
| BHI | BC/(1 μg·mL-1) | BHI | BC/(1 μg·mL-1) | BHI | BC/(1 μg·mL-1) | ||||
| EGDe | 3.488±0.116 | 22.246±0.249 | 0.205±0.0115 | 0.0842±0.00641 | 0.7125±0.00200 | 0.615±0.045 | |||
| ΔagrA | 3.348±0.196 | 16.774±0.352*** | 0.185±0.0167 | 0.1300±0.01970* | 0.7108±0.00062 | 0.566±0.005 | |||
| ΔagrB/D | 3.311±0.143 | 27.401±0.208** | 0.199±0.0129 | 0.0484±0.00171** | 0.7047±0.00960 | 0.577±0.063 | |||
| ΔagrC | 3.503±0.154 | 40.803±0.111*** | 0.196±0.0147 | 0.0646±0.00321* | 0.7005±0.00710 | 0.211±0.155* | |||
Table 3 Growth curve analysis
| 菌株 Strain | 迟缓期LPD Lag phase duration/h | 平均最大生长速率MMGR Mean maximum growth rate/(Units·h-1) | 平均最大光密度值MMOD Mean maximum optical density/Units | ||||||
|---|---|---|---|---|---|---|---|---|---|
| BHI | BC/(1 μg·mL-1) | BHI | BC/(1 μg·mL-1) | BHI | BC/(1 μg·mL-1) | ||||
| EGDe | 3.488±0.116 | 22.246±0.249 | 0.205±0.0115 | 0.0842±0.00641 | 0.7125±0.00200 | 0.615±0.045 | |||
| ΔagrA | 3.348±0.196 | 16.774±0.352*** | 0.185±0.0167 | 0.1300±0.01970* | 0.7108±0.00062 | 0.566±0.005 | |||
| ΔagrB/D | 3.311±0.143 | 27.401±0.208** | 0.199±0.0129 | 0.0484±0.00171** | 0.7047±0.00960 | 0.577±0.063 | |||
| ΔagrC | 3.503±0.154 | 40.803±0.111*** | 0.196±0.0147 | 0.0646±0.00321* | 0.7005±0.00710 | 0.211±0.155* | |||
| 菌株 Strain | 生物被膜量Biofilm mass | ||
|---|---|---|---|
| 1 d | 2 d | 3 d | |
| EGDe | 0.219±0.044 | 1.408±0.114 | 0.611±0.086 |
| ?agrA | 0.106±0.017 | 0.968±0.041 | 0.342±0.040 |
| ?agrB/D | 0.103±0.025 | 1.043±0.070 | 0.330±0.057 |
| ?agrC | 0.162±0.052 | 1.509±0.102 | 0.263±0.030 |
Table 4 Biofilm biomasses of wild type and mutant strains at 37℃
| 菌株 Strain | 生物被膜量Biofilm mass | ||
|---|---|---|---|
| 1 d | 2 d | 3 d | |
| EGDe | 0.219±0.044 | 1.408±0.114 | 0.611±0.086 |
| ?agrA | 0.106±0.017 | 0.968±0.041 | 0.342±0.040 |
| ?agrB/D | 0.103±0.025 | 1.043±0.070 | 0.330±0.057 |
| ?agrC | 0.162±0.052 | 1.509±0.102 | 0.263±0.030 |
| 菌株 Strain | 生物被膜量Biofilm mass | ||
|---|---|---|---|
| 3 d | 6 d | 9 d | |
| EGDe | 0.145±0.012 | 0.630±0.070 | 0.436±0.023 |
| ?agrA | 0.102±0.004 | 0.612±0.033 | 0.425±0.019 |
| ?agrB/D | 0.109±0.007 | 0.706±0.064 | 0.389±0.017 |
| ?agrC | 0.099±0.003 | 0.621±0.009 | 0.474±0.013 |
Table 5 Biofilm biomasses of wild type and mutant strains at 20℃
| 菌株 Strain | 生物被膜量Biofilm mass | ||
|---|---|---|---|
| 3 d | 6 d | 9 d | |
| EGDe | 0.145±0.012 | 0.630±0.070 | 0.436±0.023 |
| ?agrA | 0.102±0.004 | 0.612±0.033 | 0.425±0.019 |
| ?agrB/D | 0.109±0.007 | 0.706±0.064 | 0.389±0.017 |
| ?agrC | 0.099±0.003 | 0.621±0.009 | 0.474±0.013 |
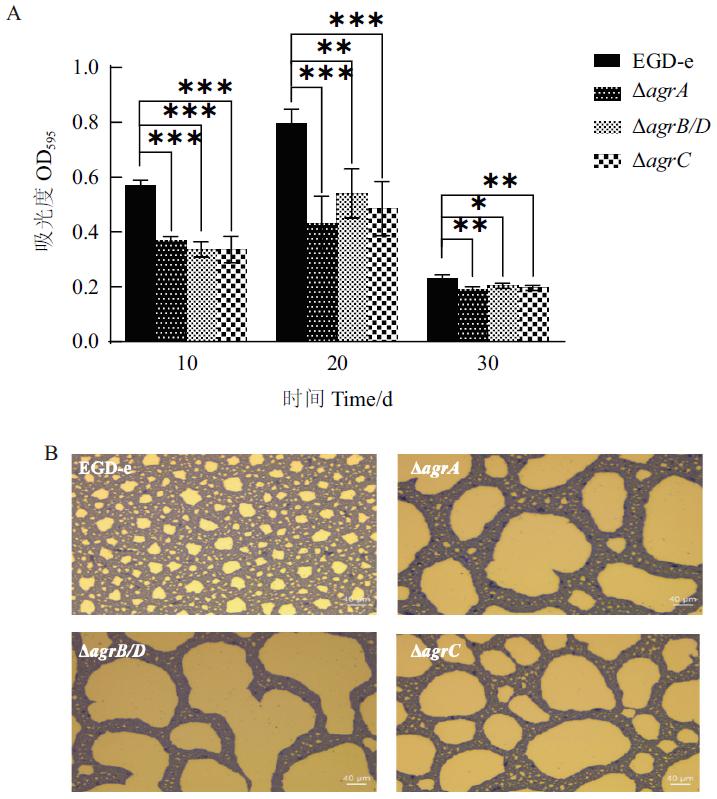
Fig. 7 Biofilm formations of L. monocytogenes strains at 4℃ 20 d A: The biofilm biomass. B: Biofilm assay by inverted microscope(incubated at 4℃ for 20 d)
| 菌株 Strain | 生物被膜量Biofilm mass | ||
|---|---|---|---|
| 10 d | 20 d | 30 d | |
| EGDe | 0.568±0.017 | 0.794±0.043 | 0.229±0.012 |
| ?agrA | 0.371±0.011 | 0.430±0.082 | 0.191±0.007 |
| ?agrB/D | 0.336±0.022 | 0.540±0.073 | 0.204±0.008 |
| ?agrC | 0.336±0.039 | 0.485±0.080 | 0.196±0.007 |
Table 6 Biofilm biomasses of wild type and mutant strains at 4℃
| 菌株 Strain | 生物被膜量Biofilm mass | ||
|---|---|---|---|
| 10 d | 20 d | 30 d | |
| EGDe | 0.568±0.017 | 0.794±0.043 | 0.229±0.012 |
| ?agrA | 0.371±0.011 | 0.430±0.082 | 0.191±0.007 |
| ?agrB/D | 0.336±0.022 | 0.540±0.073 | 0.204±0.008 |
| ?agrC | 0.336±0.039 | 0.485±0.080 | 0.196±0.007 |
| [1] | 杨丽玉, 杨诗怡, 林巍, 等. 单核细胞增生李斯特菌膜囊泡的制备及生物活性[J]. 微生物学通报, 2021, 48(1): 145-155. |
| Yang LY, Yang SY, Lin W, et al. Isolation and activity of Listeria monocytogenes-derived membrane vesicles[J]. Microbiol China, 2021, 48(1): 145-155. | |
| [2] | 耿忆敏, 任思雨, 于涛, 等. VirAB在单核细胞增生李斯特菌耐药性及生物被膜形成中的作用[J]. 微生物学通报, 2021, 48(2): 471-479. |
| Geng YM, Ren SY, Yu T, et al. Role of VirAB in antimicrobial resistance and biofilm formation of Listeria monocytogene[J]. Microbiol China, 2021, 48(2): 471-479. | |
| [3] |
Karygianni L, Ren Z, Koo H, et al. Biofilm matrixome: extracellular components in structured microbial communities[J]. Trends Microbiol, 2020, 28(8): 668-681.
doi: S0966-842X(20)30087-1 pmid: 32663461 |
| [4] |
Prescott RD, Decho AW. Flexibility and adaptability of quorum sensing in nature[J]. Trends Microbiol, 2020, 28(6): 436-444.
doi: S0966-842X(19)30319-1 pmid: 32001099 |
| [5] |
Bruger EL, Snyder DJ, Cooper VS, et al. Quorum sensing provides a molecular mechanism for evolution to tune and maintain investment in cooperation[J]. ISME J, 2021, 15(4): 1236-1247.
doi: 10.1038/s41396-020-00847-0 pmid: 33342998 |
| [6] | 刘蕾, 桂萌, 武瑞赟, 等. LuxS/AI-2型群体感应系统调控细菌生物被膜形成研究进展[J]. 食品科学, 2016, 37(19): 254-262. |
| Liu L, Gui M, Wu RY, et al. Progress in research on biofilm formation regulated by LuxS/AI-2 quorum sensing[J]. Food Sci, 2016, 37(19): 254-262. | |
| [7] |
Autret N, Raynaud C, Dubail I, et al. Identification of the agr locus of Listeria monocytogenes: role in bacterial virulence[J]. Infect Immun, 2003, 71(8): 4463-4471.
doi: 10.1128/IAI.71.8.4463-4471.2003 URL |
| [8] |
Vivant AL, Garmyn D, Gal L, et al. Survival of Listeria monocytogenes in soil requires AgrA-mediated regulation[J]. Appl Environ Microbiol, 2015, 81(15): 5073-5084.
doi: 10.1128/AEM.04134-14 URL |
| [9] | Vivant AL, Garmyn D, Gal L, et al. The Agr communication system provides a benefit to the populations of Listeria monocytogenes in soil[J]. Front Cell Infect Microbiol, 2014, 4: 160. |
| [10] |
Paspaliari DK, Mollerup MS, Kallipolitis BH, et al. Chitinase expression in Listeria monocytogenes is positively regulated by the Agr system[J]. PLoS One, 2014, 9(4): e95385.
doi: 10.1371/journal.pone.0095385 URL |
| [11] | Lee YJ, Wang C. Links between S-adenosylmethionine and Agr-based quorum sensing for biofilm development in Listeria monocytogenes EGD-E[J]. MicrobiologyOpen, 2020, 9(5): e1015. |
| [12] | Merchel Piovesan Pereira B, Tagkopoulos I. Benzalkonium chlorides: uses, regulatory status, and microbial resistance[J]. Appl Environ Microbiol, 2019, 85(13): e00377-e00319. |
| [13] |
Barber OW, Hartmann EM. Benzalkonium chloride: a systematic review of its environmental entry through wastewater treatment, potential impact, and mitigation strategies[J]. Crit Rev Environ Sci Technol, 2022, 52(15): 2691-2719.
doi: 10.1080/10643389.2021.1889284 URL |
| [14] | Kampf G. Biocidal agents used for disinfection can enhance antibiotic resistance in gram-negative species[J]. Antibiotics(Basel), 2018, 7(4): 110. |
| [15] |
Karygianni L, Ren Z, Koo H, et al. Biofilm matrixome: extracellular components in structured microbial communities[J]. Trends Microbiol, 2020, 28(8): 668-681.
doi: S0966-842X(20)30087-1 pmid: 32663461 |
| [16] |
Mazaheri T, Cervantes-Huamán BRH, Bermúdez-Capdevila M, et al. Listeria monocytogenes biofilms in the food industry: is the current hygiene program sufficient to combat the persistence of the pathogen?[J]. Microorganisms, 2021, 9(1): 181.
doi: 10.3390/microorganisms9010181 URL |
| [17] | 田翠芳, 张昭寰, 陶倩, 等. 抗生物被膜材料在食品微生物安全领域的应用研究进展[J]. 食品科学, 2022, 43(11): 265-272. |
| Tian CF, Zhang ZH, Tao Q, et al. Progress in the application of anti-biofilm materials in the field of food microbial safety[J]. Food Sci, 2022, 43(11): 265-272. | |
| [18] |
da Silva DAL, de Melo Tavares R, Camargo AC, et al. Biofilm growth by Listeria monocytogenes on stainless steel and expression of biofilm-related genes under stressing conditions[J]. World J Microbiol Biotechnol, 2021, 37(7): 119.
doi: 10.1007/s11274-021-03092-5 URL |
| [19] |
Janež N, Škrlj B, Sterniša M, et al. The role of the Listeria monocytogenes surfactome in biofilm formation[J]. Microb Biotechnol, 2021, 14(4): 1269-1281.
doi: 10.1111/1751-7915.13847 pmid: 34106516 |
| [20] | Henly EL, Dowling JAR, Maingay JB, et al. Biocide exposure induces changes in susceptibility, pathogenicity, and biofilm formation in uropathogenic Escherichia coli[J]. Antimicrob Agents Chemother, 2019, 63(3): e01892-e01818. |
| [21] |
O’May C, Ciobanu A, Lam H, et al. Tannin derived materials can block swarming motility and enhance biofilm formation in Pseudomonas aeruginosa[J]. Biofouling, 2012, 28(10): 1063-1076.
doi: 10.1080/08927014.2012.725130 URL |
| [1] | DU Dong-dong, QIAN Jing, LI Si-qi, LIU Wen-fei, WEI Xiang-li, LIU Chang-yong, LUO Rui-feng, KANG Li-chao. Whole Genome Sequencing and Analysis of Listeria monocytogenes Strain LMXJ15 [J]. Biotechnology Bulletin, 2023, 39(7): 298-306. |
| [2] | SHI Cheng-long, WANG Xi-wu, LI An-qi, QIAN Sen-he, WANG Zhou, ZHAO Shi-guang, LIU Yan, XUE Zheng-lian. Effect of ε-Polylysine on the Cell Structure and Biofilm Formation of Cronobacter sakazakii [J]. Biotechnology Bulletin, 2022, 38(9): 147-157. |
| [3] | LIU Cheng-cheng, HU Xiao-fang, FENG You-jun. Antimicrobial Resistance:Biochemical Mechanisms and Countermeasures [J]. Biotechnology Bulletin, 2022, 38(9): 4-16. |
| [4] | ZHANG Yang, CHENG Peng, LI Xiao-fen, CHEN Hong-wei. Research Progress on Anti-biofilm Peptides [J]. Biotechnology Bulletin, 2021, 37(2): 216-223. |
| [5] | LI Xiao-yan, LI Ze-qi, WANG Yu-qian, YU Jing, LIN Zhen-ping, LIN Xiang-min. Construction of Aeromonas hydrophila acrA Deficient Strain and Determination of Its Physiological Function [J]. Biotechnology Bulletin, 2020, 36(11): 63-69. |
| [6] | LIU Qian-qian, SHI Hong-wei, GUO Chang-lu, ZHANG Zhi-zhou. Composition Analysis of Prokaryotic Flora in the Marine Biofilm Correlated with Ascidian Settlement [J]. Biotechnology Bulletin, 2020, 36(11): 76-84. |
| [7] | CHANG Guo-wei, HUANG Zeng-wei, LI Zhi-de, LIANG Da-feng. Research Progress on the Development and Application of Dextranase [J]. Biotechnology Bulletin, 2019, 35(6): 196-204. |
| [8] | ZHI Wei, MA Hai-yan, QIU Yong-feng, HU Su-juan, WEI Ling-ling, ZHUAi-hua. Analysis of Antimicrobial Resistance and Resistance Genes of Salmonella from Swine [J]. Biotechnology Bulletin, 2018, 34(3): 170-176. |
| [9] | PAN Yu-rong, ZHANG Cai-li, ZHU Su-qin, ZENG Ming-yong. Inhibition of Brominated Furanone to Quorum Sensing Regulating Behaviors of Vibrio anguillarum [J]. Biotechnology Bulletin, 2017, 33(4): 231-237. |
| [10] | YAN Ning, YANG Zhi-min, SHANG Li-guo, DAI Shu-ling, ZHAN Yu-hua, LU Wei, LIN Min, YAN Yong-liang. Patterns of Biofilm Formation in Pseudomonas stutzei Under Abiotic Stresses [J]. Biotechnology Bulletin, 2017, 33(2): 172-178. |
| [11] | JIANG Xiao-bing, YU Tao, NIU Ya-bing, XU Ya-meng, SHI Lei, WANG Hai-lei. Study on the Quinolone-resistant Mechanisms of Listeria monocytogenes [J]. Biotechnology Bulletin, 2016, 32(7): 234-241. |
| [12] | ZHANG Shu-mei,XU Xiang-rong,XU Hao. Research Progress on Quorum Sensing System of Bacterial Biofilm [J]. Biotechnology Bulletin, 2016, 32(12): 19-22. |
| [13] | Zhang Lanhe, Zuo Zhengyan, Wang Xuming. Research Progress on Microbial Community Structure in Solid-phase Denitrification Systems [J]. Biotechnology Bulletin, 2015, 31(1): 39-45. |
| [14] | Liu Guishen, Yu Tao. Study on Antimicrobial Resistance and Plasmid-mediated Quinolone Resistance of Foodborne Salmonella Isolates [J]. Biotechnology Bulletin, 2014, 0(8): 202-207. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||