








Biotechnology Bulletin ›› 2023, Vol. 39 ›› Issue (4): 71-80.doi: 10.13560/j.cnki.biotech.bull.1985.2022-0641
Previous Articles Next Articles
YANG Jun-zhao( ), ZHANG Xin-rui, ZHAO Guo-zhu, ZHENG Fei(
), ZHANG Xin-rui, ZHAO Guo-zhu, ZHENG Fei( )
)
Received:2022-05-24
Online:2023-04-26
Published:2023-05-16
YANG Jun-zhao, ZHANG Xin-rui, ZHAO Guo-zhu, ZHENG Fei. Structure and Function Analysis of Novel GH5 Multi-domain Cellulase[J]. Biotechnology Bulletin, 2023, 39(4): 71-80.
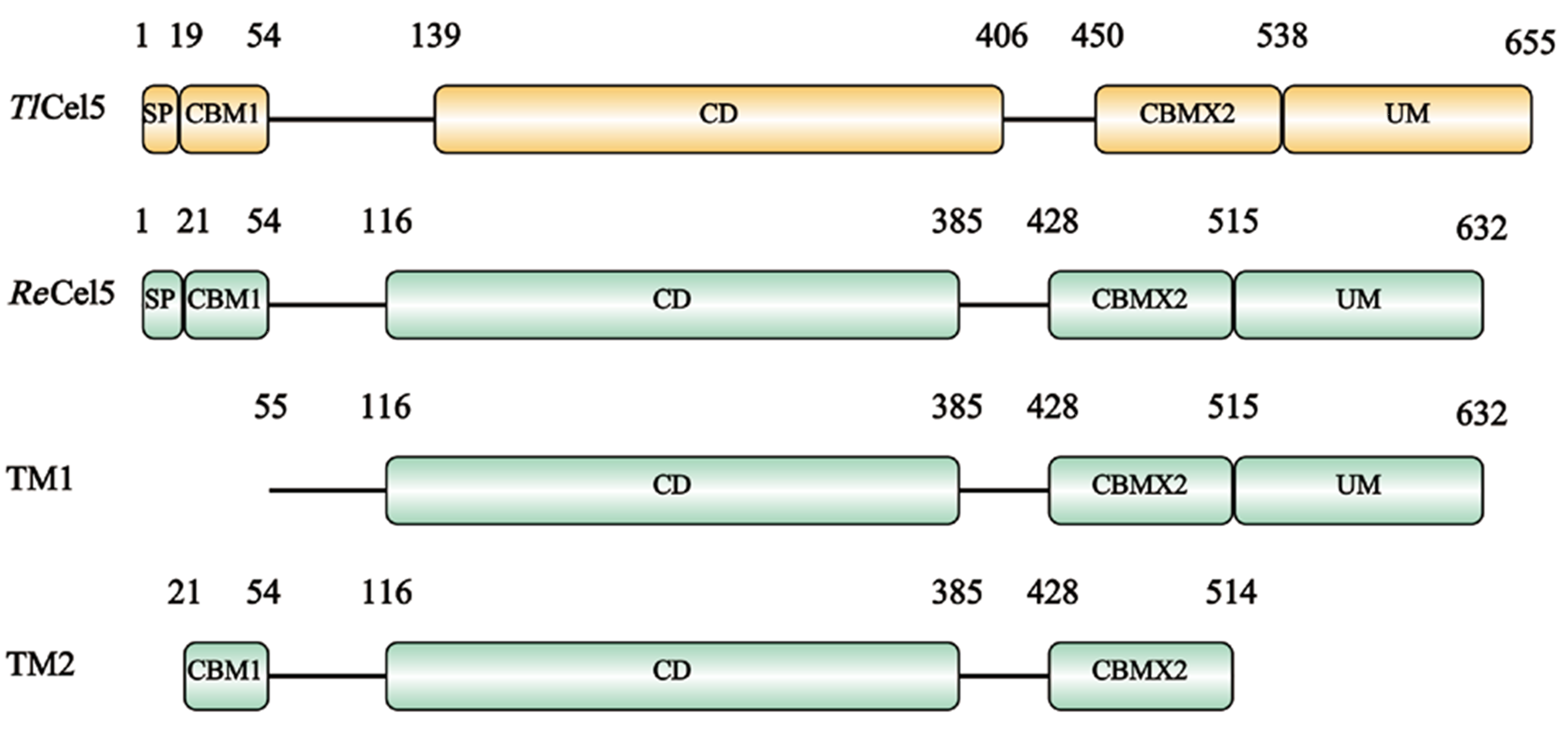
Fig. 1 Schematic diagram of protein structure encoded by TlCel5, ReCel5, TM1 and TM2 SP: Signal peptide. CBM: Carbohydrate binding module. CD: Catalysis domain. UM: Unknown module

Fig. 2 Amino acids sequence alignment in the catalytic domain of GH5 family endoglucanases The conservative residues are labeled in the arrow below the alignment. The strain source and GenBank number of the alignment sequence are H. jecorina(AEJ36301.1), R. emersonii CBS 393.64(XP_013323622.1), A. neoniger CBS 115656(XP_025478804.1), T. aurantiacus(AAL88714.2), S. opalus(ARO48344.1), G. lucidum(QDK64599.1), T. reesei(P07982.1), and G. trabeum ATCC 11539(XP_007867902.1)

Fig. 3 SDS-PAGE analysis of wild-type TlCel5, ReCel5 and mutant TM1, TM2 M: Protein molecular weight standard. A: Expression of wile-type TlCel5、ReCel5(1: purified protein of ReCel5; 2: purified protein of TlCel5). B: Expressions of mutants TM1 and TM2(3: purified protein of TM1; 4: purified protein of TM2)

Fig. 4 Enzymatic properties of wild-type TlCel5, ReCel5 and mutant TM1, TM2 A: pH-activity profile. B: pH stability. C: Temperature-activity profile. D: Thermostability at 50℃. E: Thermostability at 60℃. F: Thermostability at 70℃
| Enzyme | Vmax/(mmol·min-1·mg-1) | Km/(mg·mL-1) | kcat/(s-1) | kcat/Km(mL·mg-1·s-1) |
|---|---|---|---|---|
| TlCel5 | 217.0±20.0 | 7.5±1.0 | 247.0±22.0 | 32.8±1.5 |
| ReCel5 | 56.0±0.9 | 8.6±0.2 | 61.4±0.9 | 7.1±0.1 |
| TM1 | 68.0±0.5 | 8.4±0.1 | 70.7±0.6 | 8.4±0.1 |
| TM2 | 76.9±0.7 | 8.4±0.1 | 68.1±0.6 | 8.1±0.1 |
Table 1 Kinetics parameters of wild-type TlCel5, ReCel5 and mutants TM1, TM2
| Enzyme | Vmax/(mmol·min-1·mg-1) | Km/(mg·mL-1) | kcat/(s-1) | kcat/Km(mL·mg-1·s-1) |
|---|---|---|---|---|
| TlCel5 | 217.0±20.0 | 7.5±1.0 | 247.0±22.0 | 32.8±1.5 |
| ReCel5 | 56.0±0.9 | 8.6±0.2 | 61.4±0.9 | 7.1±0.1 |
| TM1 | 68.0±0.5 | 8.4±0.1 | 70.7±0.6 | 8.4±0.1 |
| TM2 | 76.9±0.7 | 8.4±0.1 | 68.1±0.6 | 8.1±0.1 |
| Name | Optimal pH | Optimal temperature /℃ | Source | Reference |
|---|---|---|---|---|
| TlCel5 | 3.0 | 50 | T. leycettanus | This study |
| ReCel5 | 4.0 | 70 | R. emersonii | This study |
| GlCel5A | 3.0-4.0 | 60 | Ganoderma lucidum | [ |
| TaCel5A | 6.0 | 50 | Thermoascus aurantiacusIFO9748 | [ |
| Epi3 | 6.5-7.0 | 50 | Epidinium caudatum | [ |
| PdCel5C | 4.8 | 40-50 | Penicillium decumbens 114-2 | [ |
| BaCel5 | 5.0 | 50 | Bispora antennata | [ |
| SoCel5 | 5.0 | 60 | Stegonsporium opalus | [ |
Table 2 Optimal conditions for endoglucanases of GH5 family fungi
| Name | Optimal pH | Optimal temperature /℃ | Source | Reference |
|---|---|---|---|---|
| TlCel5 | 3.0 | 50 | T. leycettanus | This study |
| ReCel5 | 4.0 | 70 | R. emersonii | This study |
| GlCel5A | 3.0-4.0 | 60 | Ganoderma lucidum | [ |
| TaCel5A | 6.0 | 50 | Thermoascus aurantiacusIFO9748 | [ |
| Epi3 | 6.5-7.0 | 50 | Epidinium caudatum | [ |
| PdCel5C | 4.8 | 40-50 | Penicillium decumbens 114-2 | [ |
| BaCel5 | 5.0 | 50 | Bispora antennata | [ |
| SoCel5 | 5.0 | 60 | Stegonsporium opalus | [ |

Fig. 6 Sequence alignment of CBM1 domain of cellulose The cellulase name, source strain and GenBank number of the alignment sequence are ReCel5(R. emersonii CBS 393.64, XP_013323622.1), TrCel7A(T. reesei, CAH10320.1), TrCel7B(T. reesei, AAA34212.1), PdCel5C(P. decumbens, JQ319040.1), TrCel5C(T. reesei, AAA34213.1), ApCel5A(Aureobasidium pullulans, AEM23896.1)
| [1] | 赵琪, 李亚兰, 陈子欣, 等. 纤维素酶应用研究的最新进展[J]. 广州化工, 2014, 42(6): 21-23. |
| Zhao Q, Li YL, Chen ZX, et al. The recent application progress cellulase[J]. Guangzhou Chem Ind, 2014, 42(6): 21-23. | |
| [2] |
Xin DL, Blossom BM, Lu XY, et al. Improving cellulases hydrolytic action: an expanded role for electron donors of lytic polysaccharide monooxygenases in cellulose saccharification[J]. Bioresour Technol, 2022, 346: 126662.
doi: 10.1016/j.biortech.2021.126662 URL |
| [3] |
Sidar A, Albuquerque ED, Voshol GP, et al. Carbohydrate binding modules: diversity of domain architecture in amylases and cellulases from filamentous microorganisms[J]. Front Bioeng Biotechnol, 2020, 8: 871.
doi: 10.3389/fbioe.2020.00871 URL |
| [4] |
Hu YM, Li HN, Ran QP, et al. Effect of carbohydrate binding modules alterations on catalytic activity of glycoside hydrolase family 6 exoglucanase from Chaetomium thermophilum to cellulose[J]. Int J Biol Macromol, 2021, 191: 222-229.
doi: 10.1016/j.ijbiomac.2021.09.002 URL |
| [5] |
Shibata N, Suetsugu M, Kakeshita H, et al. A novel GH10 xylanase from Penicillium sp. accelerates saccharification of alkaline-pretreated bagasse by an enzyme from recombinant Trichoderma reesei expressing Aspergillus β-glucosidase[J]. Biotechnol Biofuels, 2017, 10: 278.
doi: 10.1186/s13068-017-0970-2 pmid: 29201142 |
| [6] |
Thongekkaew J, Ikeda H, Masaki K, et al. Fusion of cellulose binding domain from Trichoderma reesei CBHI to Cryptococcus sp. S-2 cellulase enhances its binding affinity and its cellulolytic activity to insoluble cellulosic substrates[J]. Enzyme Microb Technol, 2013, 52(4/5): 241-246.
doi: 10.1016/j.enzmictec.2013.02.002 URL |
| [7] | 孔海洋, 蒋肖, 王苑, 等. β-甘露聚糖酶Man5A和木聚糖酶Tlxyn11B的融合表达[J]. 生物工程学报, 2020, 36(9): 1849-1858. |
| Kong HY, Jiang X, Wang Y, et al. Fusion expression of β-mannanase Man5A and xylanase Tlxyn11B in Pichia pastoris[J]. Chin J Biotechnol, 2020, 36(9): 1849-1858. | |
| [8] |
Liu GZ, Li Q, Shang N, et al. Functional and structural analyses of a 1, 4-β-endoglucanase from Ganoderma lucidum[J]. Enzyme Microb Technol, 2016, 86: 67-74.
doi: 10.1016/j.enzmictec.2016.01.013 URL |
| [9] |
Hong J, Tamaki H, Yamamoto K, et al. Cloning of a gene encoding a thermo-stable endo-beta-1, 4-glucanase from Thermoascus aurantiacus and its expression in yeast[J]. Biotechnol Lett, 2003, 25(8): 657-661.
doi: 10.1023/A:1023072311980 URL |
| [10] |
Takenak A, D’Silva CG, Kudo H, et al. Molecular cloning, expression, and characterization of an endo-beta-1, 4-glucanase cDNA from Epidinium caudatum1[J]. J Gen Appl Microbiol, 1999, 45(2): 57-61.
pmid: 12501388 |
| [11] |
Liu GD, Qin YQ, Hu YB, et al. An endo-1, 4-β-glucanase PdCel5C from cellulolytic fungus Penicillium decumbens with distinctive domain composition and hydrolysis product profile[J]. Enzyme Microb Technol, 2013, 52(3): 190-195.
doi: 10.1016/j.enzmictec.2012.12.009 URL |
| [12] |
Zheng F, Huang HQ, Wang XY, et al. Improvement of the catalytic performance of a Bispora antennata cellulase by replacing the N-terminal semi-barrel structure[J]. Bioresour Technol, 2016, 218: 279-285.
doi: 10.1016/j.biortech.2016.06.094 URL |
| [13] | Zheng F, Vermaas JV, Zheng J, et al. Activity and thermostability of GH5 endoglucanase chimeras from mesophilic and thermophilic parents[J]. Appl Environ Microbiol, 2019, 85(5): e02079-e02018. |
| [14] |
Inoue H, Kishishita S, Kumagai A, et al. Contribution of a family 1 carbohydrate-binding module in thermostable glycoside hydrolase 10 xylanase from Talaromyces cellulolyticus toward synergistic enzymatic hydrolysis of lignocellulose[J]. Biotechnol Biofuels, 2015, 8: 77.
doi: 10.1186/s13068-015-0259-2 URL |
| [15] |
Várnai A, Siika-Aho M, Viikari L. Carbohydrate-binding modules(CBMs)revisited: reduced amount of water counterbalances the need for CBMs[J]. Biotechnol Biofuels, 2013, 6(1): 30.
doi: 10.1186/1754-6834-6-30 pmid: 23442543 |
| [16] |
Ståhlberg J, Johansson G, Pettersson G. A binding-site-deficient, catalytically active, core protein of endoglucanase III from the culture filtrate of Trichoderma reesei[J]. Eur J Biochem, 1988, 173(1): 179-183.
pmid: 3356188 |
| [17] |
Kraulis J, Clore GM, Nilges M, et al. Determination of the three-dimensional solution structure of the C-terminal domain of cellobiohydrolase I from Trichoderma reesei. A study using nuclear magnetic resonance and hybrid distance geometry-dynamical simulated annealing[J]. Biochemistry, 1989, 28(18): 7241-7257.
doi: 10.1021/bi00444a016 pmid: 2554967 |
| [18] |
Hervé C, Rogowski A, Blake AW, et al. Carbohydrate-binding modules promote the enzymatic deconstruction of intact plant cell walls by targeting and proximity effects[J]. Proc Natl Acad Sci USA, 2010, 107(34): 15293-15298.
doi: 10.1073/pnas.1005732107 pmid: 20696902 |
| [19] |
Bolam DN, Ciruela A, Mcqueen-Mason S, et al. Pseudomonas cellulose-binding domains mediate their effects by increasing enzyme substrate proximity[J]. Biochem J, 1998, 331(Pt 3): 775-781.
doi: 10.1042/bj3310775 URL |
| [20] |
Aich S, Datta S. Engineering of a highly thermostable endoglucanase from the GH7 family of Bipolaris sorokiniana for higher catalytic efficiency[J]. Appl Microbiol Biotechnol, 2020, 104(9): 3935-3945.
doi: 10.1007/s00253-020-10515-0 |
| [21] |
Linder M, Lindeberg G, Reinikainen T, et al. The difference in affinity between two fungal cellulose-binding domains is dominated by a single amino acid substitution[J]. FEBS Lett, 1995, 372(1): 96-98.
pmid: 7556652 |
| [22] |
Arola S, Linder MB. Binding of cellulose binding modules reveal differences between cellulose substrates[J]. Sci Rep, 2016, 6: 35358.
doi: 10.1038/srep35358 pmid: 27748440 |
| [23] |
Nishijima H, Nozaki K, Mizuno M, et al. Extra tyrosine in the carbohydrate-binding module of Irpex lacteus Xyn10B enhances its cellulose-binding ability[J]. Biosci Biotechnol Biochem, 2015, 79(5): 738-746.
doi: 10.1080/09168451.2014.996203 URL |
| [1] | WANG Yu-chen, DING Zun-dan, GUAN Fei-fei, TIAN Jian, LIU Guo-an, WU Ning-feng. Identification of the Thermostable Laccase Gene ba4 and Characterization of Its Enzymatic Properties [J]. Biotechnology Bulletin, 2022, 38(8): 252-260. |
| [2] | FU Qiao, LIN Qi-lan, XUE Qiang, XIONG Hai-rong, WANG Ya-wei. Effects of CBM41 N-terminal Truncation on the Enzymological Properties of the Pullulanase from Bacillus subtilis 168 [J]. Biotechnology Bulletin, 2022, 38(6): 245-251. |
| [3] | WANG Xiao-tao, ZOU Hang, WU Yi, XIANG Shen-wei, LV Hua, LIU Chao-lan, LIN Jia-fu, WANG Xin-rong, CHU Yi-wen, SONG Tao. Heterologous Expression and Enzymatic Properties Analysis of Novel β-agarase Aga2 from Paraglaciecola hydrolytica [J]. Biotechnology Bulletin, 2022, 38(11): 258-268. |
| [4] | ZHANG Yao-xin, WANG Liang-jie, ZHENG Wen, XU Han-qin, ZHENG Lian, ZHONG Jing. Study on Enzyme Production of a Chitinase-producing Strain Achromobacter sp. ZWW8 by Fermentation and Its Enzymatic Characterization [J]. Biotechnology Bulletin, 2021, 37(4): 96-106. |
| [5] | WANG Hui-lan, WU Jin-yong, CHEN Xiang-song, YUAN Li-xia, ZHU Wei-wei, YAO Jian-ming. Immobilization of N-acetylneuraminic Acid Aldolaseand Properties of the Immobilized Enzyme [J]. Biotechnology Bulletin, 2020, 36(6): 165-173. |
| [6] | LI Cheng-ming, WANG Jia-yi, LU Lei. Extracellular Expression of Laccase Gene from Bacillus pumilus LC01 in Pichia pastoris and Characterization of the Recombinant Laccase [J]. Biotechnology Bulletin, 2018, 34(3): 185-193. |
| [7] | ZHAO Shu-mei, WANG Lin-hui, TANG Jia-qi, ZHAO Jing-yi. Cloning,Expression and Characterization of the Esterase Gene estZ from Vibrio alginolyticus [J]. Biotechnology Bulletin, 2017, 33(6): 190-196. |
| [8] | Liu Song, Lu Xinyao, Zhou Jingwen, Du Guocheng, Chen Jian. Research Advance on the Structure, Molecular Modification, and Fermentation of Lipoxygenases [J]. Biotechnology Bulletin, 2015, 31(12): 34-41. |
| [9] | Jia Yang, Shi Yanhua, Ren Lei, Qiao Cheng, Wang Junhuan, Yan Yanchun. Cloning and Characterization of mpd and ophc2 from an Organophosphorus Pesticide-degrading Bacteria YC-YH1 [J]. Biotechnology Bulletin, 2015, 31(11): 228-235. |
| [10] | Zhu Licheng, Luo Hui, Ren Han. Progress in Structure Function and Antifungal Mechanism of Plant Defensins [J]. Biotechnology Bulletin, 2014, 0(3): 9-14. |
| [11] | Xing Zhaobin, Wu Peng, Li Feifei, Liu Yan, Zhou Mi, Xiu Leshan. Cloning and Expression Analysis of Cytochrome P450 Gene in Eleutherococcus senticosus [J]. Biotechnology Bulletin, 2014, 0(1): 112-115. |
| [12] | Liu Yanjie, Li Haitao, Liu Rongmei, Gao Jiguo. Bt New Gene sip Cloning, Expression and Bioinformatics Analysis [J]. Biotechnology Bulletin, 2012, 0(12): 101-105. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||