








Biotechnology Bulletin ›› 2023, Vol. 39 ›› Issue (5): 224-232.doi: 10.13560/j.cnki.biotech.bull.1985.2022-1285
Previous Articles Next Articles
ZHAI Ying1( ), LI Ming-yang1, ZHANG Jun2, ZHAO Xu3, YU Hai-wei1, LI Shan-shan1, ZHAO Yan1, ZHANG Mei-juan1, SUN Tian-guo1
), LI Ming-yang1, ZHANG Jun2, ZHAO Xu3, YU Hai-wei1, LI Shan-shan1, ZHAO Yan1, ZHANG Mei-juan1, SUN Tian-guo1
Received:2022-10-18
Online:2023-05-26
Published:2023-06-08
ZHAI Ying, LI Ming-yang, ZHANG Jun, ZHAO Xu, YU Hai-wei, LI Shan-shan, ZHAO Yan, ZHANG Mei-juan, SUN Tian-guo. Heterologous Expression of Soybean Transcription Factor GmNF-YA19 Improves Drought Resistance of Transgenic Tobacco[J]. Biotechnology Bulletin, 2023, 39(5): 224-232.
| 基因(GenBank登录号)Genes(GenBank accession number) | 正向引物Forward primer(5'-3') | 反向引物Reverse primer(5'-3') |
|---|---|---|
| Gmβ-tubulin(GMU12286) | GGAAGGCTTTCTTGCATTGGTA | AGTGGCATCCTGGTACTGC |
| GmNF-YA19(NM001253093) | AGCCATTCTTGTCGAATCATACTG | CCTGTGGTCCATAAGCAACTATTG |
| NtActin(AB158612) | TTGCTGGTCGTGATCTTACTGATTG | CAGTCTCCAACTCTTGCTCATAGTC |
| NtABA2(EU123520) | TCTCCAAACTTTCCTACTCGCTTTC | CACTGCTTGACGCTCTTCCTTC |
| NtNCED3-1(JX101472) | CCCAAATGTGTTCAAGGCGTTTAC | TGCCGTCACCGTCAAAGAAATG |
| NtNCED3-2(JX101473) | ATATGGGAAGAAATCGCCTGATGTC | ACAACTTGCTGGTCGGGAATTAC |
| NtCAT1(U93244) | CCTCGTGGTTTTGCTGTC | GGGATTTAGGATTTGGCTT |
| NtP5CS(HM854026) | GACACGGACTGATGGAAGATTAG | TTCATAGCCTTGCGAGTTAAGC |
Table 1 Primer sequences for RT-qPCR
| 基因(GenBank登录号)Genes(GenBank accession number) | 正向引物Forward primer(5'-3') | 反向引物Reverse primer(5'-3') |
|---|---|---|
| Gmβ-tubulin(GMU12286) | GGAAGGCTTTCTTGCATTGGTA | AGTGGCATCCTGGTACTGC |
| GmNF-YA19(NM001253093) | AGCCATTCTTGTCGAATCATACTG | CCTGTGGTCCATAAGCAACTATTG |
| NtActin(AB158612) | TTGCTGGTCGTGATCTTACTGATTG | CAGTCTCCAACTCTTGCTCATAGTC |
| NtABA2(EU123520) | TCTCCAAACTTTCCTACTCGCTTTC | CACTGCTTGACGCTCTTCCTTC |
| NtNCED3-1(JX101472) | CCCAAATGTGTTCAAGGCGTTTAC | TGCCGTCACCGTCAAAGAAATG |
| NtNCED3-2(JX101473) | ATATGGGAAGAAATCGCCTGATGTC | ACAACTTGCTGGTCGGGAATTAC |
| NtCAT1(U93244) | CCTCGTGGTTTTGCTGTC | GGGATTTAGGATTTGGCTT |
| NtP5CS(HM854026) | GACACGGACTGATGGAAGATTAG | TTCATAGCCTTGCGAGTTAAGC |

Fig. 1 Expressions of GmNF-YA19 under drought, salt, cold and ABA treatments Different letters indicate significant difference(P <0.05). The same below

Fig. 2 Nucleotide and protein amino acid sequences of GmNF-YA19 The shadow is the CBF domain. The underline is NF-YB/NF-YC binding subdomain. The bold one is the DNA binding subdomain. * is the termination codon
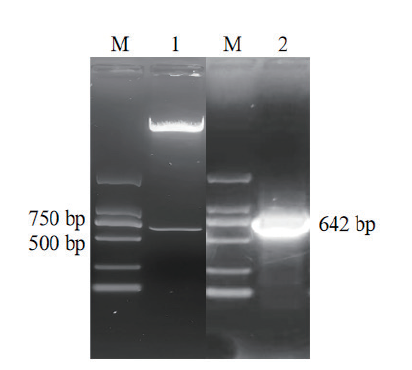
Fig. 4 Construction of GmNF-YA19 plant expression vector and Agrobacterium tumefaciens transformation M: DL2000 marker. 1: Double enzyme digestion of pRI101-GmNF-YA19 plasmid.2: PCR of Agrobacterium tumefaciens
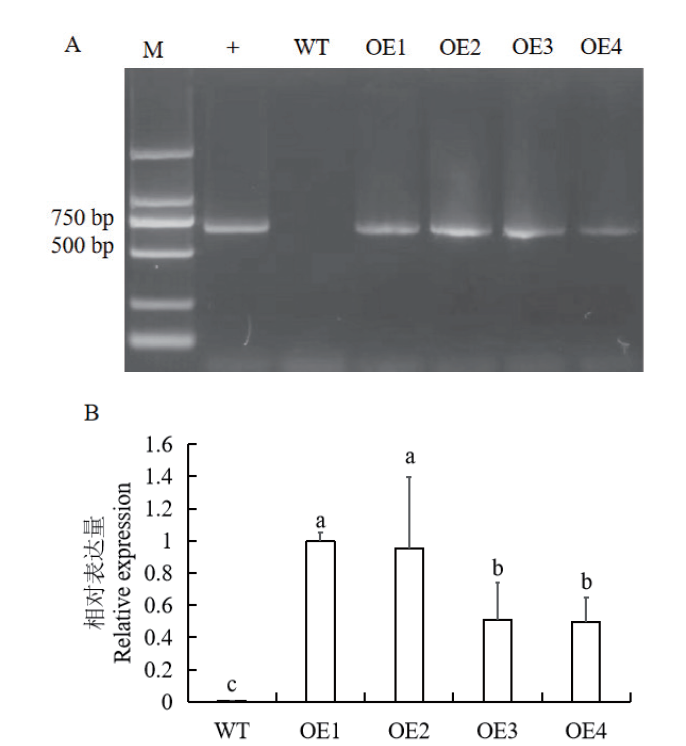
Fig. 5 Genomic DNA PCR detection (A) and RT-qPCR detection (B) of GmNF-YA19 transgenic tobacco M: DL2000 marker. +: pRI101-GmNF-YA19 plasmid. WT: Wild-type tobacco. OE1-OE4: GmNF-YA19 transgenic tobacco plants

Fig. 6 Phenotype of GmNF-YA19 transgenic tobacco plants under drought and rehydration treatment A: Before stop watering. B: Stop watering for 10 d. C: Stop watering for 18 d. D: Rehydration 3 d. WT: Wild-type tobacco. OE1-OE3: GmNF-YA19 transgenic tobacco plants

Fig. 7 Electrolyte leakage, malondialdehyde contents, soluble carbohydrate contents and proline contents in GmNF-YA19 transgenic tobacco plants under drought stress WT: Wild-type tobacco; OE1-OE3: GmNF-YA19 transgenic tobacco plants
| [1] |
Zhu JK. Salt and drought stress signal transduction in plants[J]. Annu Rev Plant Biol, 2002, 53: 247-273.
doi: 10.1146/arplant.2002.53.issue-1 URL |
| [2] |
Xu WY, Tang WS, Wang CX, et al. SiMYB56 confers drought stress tolerance in transgenic rice by regulating lignin biosynthesis and ABA signaling pathway[J]. Front Plant Sci, 2020, 11: 785.
doi: 10.3389/fpls.2020.00785 pmid: 32625221 |
| [3] |
Lu X, Zhang XF, Duan H, et al. Three stress-responsive NAC transcription factors from Populus euphratica differentially regulate salt and drought tolerance in transgenic plants[J]. Physiol Plant, 2018, 162(1): 73-97.
doi: 10.1111/ppl.2018.162.issue-1 URL |
| [4] |
Wu J, Zhang J, Li X, et al. An overexpression of the AP2/ERF transcription factor from Iris typhifolia in Arabidopsis thaliana confers tolerance to salt stress[J]. Biol Plant, 2019, 63: 776-784.
doi: 10.32615/bp.2019.082 URL |
| [5] |
Gao HM, Wang YF, Xu P, et al. Overexpression of a WRKY transcription factor TaWRKY2 enhances drought stress tolerance in transgenic wheat[J]. Front Plant Sci, 2018, 9: 997.
doi: 10.3389/fpls.2018.00997 URL |
| [6] |
Yao PF, Li CL, Zhao XR, et al. Overexpression of a Tartary buckwheat gene, FtbHLH3, enhances drought/oxidative stress tolerance in transgenic Arabidopsis[J]. Front Plant Sci, 2017, 8: 625.
doi: 10.3389/fpls.2017.00625 URL |
| [7] |
Mantovani R. The molecular biology of the CCAAT-binding factor NF-Y[J]. Gene, 1999, 239(1): 15-27.
doi: 10.1016/s0378-1119(99)00368-6 pmid: 10571030 |
| [8] | 徐妙云, 朱佳旭, 张敏, 等. 植物miR169/NF-YA调控模块研究进展[J]. 遗传, 2016, 38(8): 700-706. |
| Xu MY, Zhu JX, Zhang M, et al. Advances on plant miR169/NF-YA regulation modules[J]. Hereditas, 2016, 38(8): 700-706. | |
| [9] |
Li WX, Oono Y, Zhu JH, et al. The Arabidopsis NFYA5 transcription factor is regulated transcriptionally and posttranscriptionally to promote drought resistance[J]. Plant Cell, 2008, 20(8): 2238-2251.
doi: 10.1105/tpc.108.059444 URL |
| [10] |
Gusmaroli G, Tonelli C, Mantovani R. Regulation of novel members of the Arabidopsis thaliana CCAAT-binding nuclear factor Y subunits[J]. Gene, 2002, 283(1/2): 41-48.
doi: 10.1016/S0378-1119(01)00833-2 URL |
| [11] | Feng ZJ, He GH, Zheng WJ, et al. Foxtail millet NF-Y families: genome-wide survey and evolution analyses identified two functional genes important in abiotic stresses[J]. Front Plant Sci, 2015, 6: 1142. |
| [12] |
Li J, Duan YJ, Sun NL, et al. The miR169n-NF-YA8 regulation module involved in drought resistance in Brassica napus L[J]. Plant Sci, 2021, 313: 111062.
doi: 10.1016/j.plantsci.2021.111062 URL |
| [13] |
Su HH, Cao YY, Ku LX, et al. Dual functions of ZmNF-YA3 in photoperiod-dependent flowering and abiotic stress responses in maize[J]. J Exp Bot, 2018, 69(21): 5177-5189.
doi: 10.1093/jxb/ery299 pmid: 30137393 |
| [14] | 刘芳兵. 狗牙根CdNF-YA23基因响应盐胁迫的功能解析[D]. 武汉: 中国科学院大学(中国科学院武汉植物园), 2020. |
| Liu FB. Functional analysis of CdNF-YA23 from bermudagrass in response to salt stress[D]. Wuhan: Wuhan Botanical Garden, Chinese Academy of Sciences, 2020. | |
| [15] |
Lian CL, Li Q, Yao K, et al. Populus trichocarpa PtNF-YA9, A multifunctional transcription factor, regulates seed germination, abiotic stress, plant growth and development in Arabidopsis[J]. Front Plant Sci, 2018, 9: 954.
doi: 10.3389/fpls.2018.00954 URL |
| [16] |
Quach TN, Nguyen HTM, Valliyodan B, et al. Genome-wide expression analysis of soybean NF-Y genes reveals potential function in development and drought response[J]. Mol Genet Genomics, 2015, 290(3): 1095-1115.
doi: 10.1007/s00438-014-0978-2 pmid: 25542200 |
| [17] |
Ni ZY, Hu Z, Jiang QY, et al. GmNFYA3, a target gene of miR169, is a positive regulator of plant tolerance to drought stress[J]. Plant Mol Biol, 2013, 82(1/2): 113-129.
doi: 10.1007/s11103-013-0040-5 URL |
| [18] |
Ma XJ, Yu TF, Li XH, et al. Overexpression of GmNFYA5 confers drought tolerance to transgenic Arabidopsis and soybean plants[J]. BMC Plant Biol, 2020, 20(1): 123.
doi: 10.1186/s12870-020-02337-z |
| [19] |
Yu YH, Bai YC, Wang P, et al. Soybean nuclear factor YA10 positively regulates drought resistance in transgenic Arabidopsis thaliana[J]. Environ Exp Bot, 2020, 180: 104249.
doi: 10.1016/j.envexpbot.2020.104249 URL |
| [20] |
Qiu S, Zhang J, He JQ, et al. Overexpression of GmGolS2-1, a soybean galactinol synthase gene, enhances transgenic tobacco drought tolerance[J]. Plant Cell Tissue Organ Cult, 2020, 143(3): 507-516.
doi: 10.1007/s11240-020-01936-w |
| [21] |
Hoekema A, Hirsch PR, Hooykaas PJJ, et al. A binary plant vector strategy based on separation of vir- and T-region of the Agrobacterium tumefaciens Ti-plasmid[J]. Nature, 1983, 303(5913): 179-180.
doi: 10.1038/303179a0 |
| [22] | 李敏, 于太飞, 徐兆师, 等. 大豆转录因子基因GmNF-YCa可提高转基因拟南芥渗透胁迫的耐性[J]. 作物学报, 2017, 43(8): 1161-1169. |
|
Li M, Yu TF, Xu ZS, et al. Soybean transcription factor gene GmNF-YCa enhances osmotic stress tolerance of transgenic Arabidopsis[J]. Acta Agron Sin, 2017, 43(8): 1161-1169.
doi: 10.3724/SP.J.1006.2017.01161 URL |
|
| [23] |
O'Conner S, Zheng WG, Qi MS, et al. GmNF-YC4-2 increases protein, exhibits broad disease resistance and expedites maturity in soybean[J]. Int J Mol Sci, 2021, 22(7): 3586.
doi: 10.3390/ijms22073586 URL |
| [24] |
Mallano AI, Li WB, Tabys D, et al. The soybean GmNFY-B1 transcription factor positively regulates flowering in transgenic Arabidopsis[J]. Mol Biol Rep, 2021, 48(2): 1589-1599.
doi: 10.1007/s11033-021-06164-9 pmid: 33512627 |
| [25] |
Lee DK, Kim HI, Jang G, et al. The NF-YA transcription factor OsNF-YA7 confers drought stress tolerance of rice in an abscisic acid independent manner[J]. Plant Sci, 2015, 241: 199-210.
doi: 10.1016/j.plantsci.2015.10.006 URL |
| [26] |
黄锁, 胡利芹, 徐东北, 等. 谷子转录因子SiNF-YA5通过ABA非依赖途径提高转基因拟南芥耐盐性[J]. 作物学报, 2016, 42(12): 1787-1797.
doi: 10.3724/SP.J.1006.2016.01787 |
|
Huang S, Hu LQ, Xu DB, et al. Transcription factor SiNF-YA5 from foxtail millet(Setaria italica)conferred tolerance to high-salt stress through ABA-independent pathway in transgenic Arabidopsis[J]. Acta Agron Sin, 2016, 42(12): 1787-1797.
doi: 10.3724/SP.J.1006.2016.01787 URL |
|
| [27] |
Leyva-González MA, Ibarra-Laclette E, Cruz-Ramírez A, et al. Functional and transcriptome analysis reveals an acclimatization strategy for abiotic stress tolerance mediated by Arabidopsis NF-YA family members[J]. PLoS One, 2012, 7(10): e48138.
doi: 10.1371/journal.pone.0048138 URL |
| [28] |
Liu JX, Howell SH. bZIP28 and NF-Y transcription factors are activated by ER stress and assemble into a transcriptional complex to regulate stress response genes in Arabidopsis[J]. Plant Cell, 2010, 22(3): 782-796.
doi: 10.1105/tpc.109.072173 URL |
| [29] |
Alam MM, Tanaka T, Nakamura H, et al. Overexpression of a rice heme activator protein gene(OsHAP2E)confers resistance to pathogens, salinity and drought, and increases photosynthesis and tiller number[J]. Plant Biotechnol J, 2015, 13(1): 85-96.
doi: 10.1111/pbi.2014.13.issue-1 URL |
| [30] |
Frey A, Audran C, Marin E, et al. Engineering seed dormancy by the modification of Zeaxanthin epoxidase gene expression[J]. Plant Mol Biol, 1999, 39(6): 1267-1274.
pmid: 10380812 |
| [31] |
Jia YY, Liu JL, Bai ZQ, et al. Cloning and functional characterization of the SmNCED3 in Salvia miltiorrhiza[J]. Acta Physiol Plant, 2018, 40(7): 1-9.
doi: 10.1007/s11738-017-2577-4 |
| [32] | 李大红, 刘宏伟, 秦兰娟, 等. 拟南芥P5CS1基因转化羽衣甘蓝增强耐盐性分析[J]. 南方农业学报, 2017, 48(5): 768-773. |
| Li DH, Liu HW, Qin LJ, et al. Salt tolerance strengthening of Brassica oleracea var. acephala f. tricolor genetically modified by P5CS1 gene from Arabidopsis thaliana[J]. J South Agric, 2017, 48(5): 768-773. |
| [1] | WANG Shuai, FENG Yu-mei, BAI Miao, DU Wei-jun, YUE Ai-qin. Functional Analysis of Soybean Gene GmHMGR Responding to Exogenous Hormones and Abiotic Stresses [J]. Biotechnology Bulletin, 2023, 39(7): 131-142. |
| [2] | LI Wen-chen, LIU Xin, KANG Yue, LI Wei, QI Ze-zheng, YU Lu, WANG Fang. Optimization and Application of Tobacco Rattle Virus-induced Gene Silencing System in Soybean [J]. Biotechnology Bulletin, 2023, 39(7): 143-150. |
| [3] | ZHAO Xue-ting, GAO Li-yan, WANG Jun-gang, SHEN Qing-qing, ZHANG Shu-zhen, LI Fu-sheng. Cloning and Expression of AP2/ERF Transcription Factor Gene ShERF3 in Sugarcane and Subcellular Localization of Its Encoded Protein [J]. Biotechnology Bulletin, 2023, 39(6): 208-216. |
| [4] | LI Yuan-hong, GUO Yu-hao, CAO Yan, ZHU Zhen-zhou, WANG Fei-fei. Research Progress in the Microalgal Growth and Accumulation of Target Products Regulated by Exogenous Phytohormone [J]. Biotechnology Bulletin, 2023, 39(6): 61-72. |
| [5] | FENG Shan-shan, WANG Lu, ZHOU Yi, WANG You-ping, FANG Yu-jie. Research Progresses on WOX Family Genes in Regulating Plant Development and Abiotic Stress Response [J]. Biotechnology Bulletin, 2023, 39(5): 1-13. |
| [6] | HOU Xiao-yuan, CHE Zheng-zheng, LI Heng-jing, DU Chong-yu, XU Qian, WANG Qun-qing. Construction of the Soybean Membrane System cDNA Library and Interaction Proteins Screening for Effector PsAvr3a [J]. Biotechnology Bulletin, 2023, 39(4): 268-276. |
| [7] | YANG Chun-hong, DONG Lu, CHEN Lin, SONG Li. Characterization of Soybean VAS1 Gene Family and Its Involvement in Lateral Root Development [J]. Biotechnology Bulletin, 2023, 39(3): 133-142. |
| [8] | CHEN Yi-bo, YANG Wan-ming, YUE Ai-qin, WANG Li-xiang, DU Wei-jun, WANG Min. Construction of Soybean Genetic Map Based on SLAF Markers and QTL Mapping Analysis of Salt Tolerance at Seedling Stage [J]. Biotechnology Bulletin, 2023, 39(2): 70-79. |
| [9] | MIAO Shu-nan, GAO Yu, LI Xin-ru, CAI Gui-ping, ZHANG Fei, XUE Jin-ai, JI Chun-li, LI Run-zhi. Functional Analysis of Soybean GmPDAT1 Genes in the Oil Biosynthesis and Response to Abiotic Stresses [J]. Biotechnology Bulletin, 2023, 39(2): 96-106. |
| [10] | XU Rui, ZHU Ying-fang. The Key Roles of Mediator Complex in Plant Responses to Abiotic Stress [J]. Biotechnology Bulletin, 2023, 39(11): 54-60. |
| [11] | CHEN Guang-xia, LI Xiu-jie, JIANG Xi-long, SHAN Lei, ZHANG Zhi-chang, LI Bo. Research Progress in Plant Small Signaling Peptides Involved in Abiotic Stress Response [J]. Biotechnology Bulletin, 2023, 39(11): 61-73. |
| [12] | HAN Fang-ying, HU Xin, WANG Nan-nan, XIE Yu-hong, WANG Xiao-yan, ZHU Qiang. Research Progress in Response of DREBs to Abiotic Stress in Plant [J]. Biotechnology Bulletin, 2023, 39(11): 86-98. |
| [13] | SUN Yu-tong, LIU De-shuai, QI Xun, FENG Mei, HUANG Xu-zheng, YAO Wen-kong. Advances in Jasmonic Acid Regulating Plant Growth and Development as Well as Stress [J]. Biotechnology Bulletin, 2023, 39(11): 99-109. |
| [14] | GE Wen-dong, WANG Teng-hui, MA Tian-yi, FAN Zhen-yu, WANG Yu-shu. Genome-wide Identification of the PRX Gene Family in Cabbage(Brassica oleracea L. var. capitata)and Expression Analysis Under Abiotic Stress [J]. Biotechnology Bulletin, 2023, 39(11): 252-260. |
| [15] | YANG Xu-yan, ZHAO Shuang, MA Tian-yi, BAI Yu, WANG Yu-shu. Cloning of Three Cabbage WRKY Genes and Their Expressions in Response to Abiotic Stress [J]. Biotechnology Bulletin, 2023, 39(11): 261-269. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||