








Biotechnology Bulletin ›› 2023, Vol. 39 ›› Issue (5): 306-313.doi: 10.13560/j.cnki.biotech.bull.1985.2022-0962
Previous Articles Next Articles
CHEN Xiao-meng( ), ZHANG Xue-jing, ZHANG Huan, ZHANG Bao-jiang, SU Yan(
), ZHANG Xue-jing, ZHANG Huan, ZHANG Bao-jiang, SU Yan( )
)
Received:2022-08-04
Online:2023-05-26
Published:2023-06-08
Contact:
SU Yan
E-mail:1292236360@qq.com;2006au@qq.com
CHEN Xiao-meng, ZHANG Xue-jing, ZHANG Huan, ZHANG Bao-jiang, SU Yan. Prokaryotic Expression of Recombinant Bovine Mastitis Staphylococcus aureus GapC Protein and Identification of Its B-cell Epitopes[J]. Biotechnology Bulletin, 2023, 39(5): 306-313.
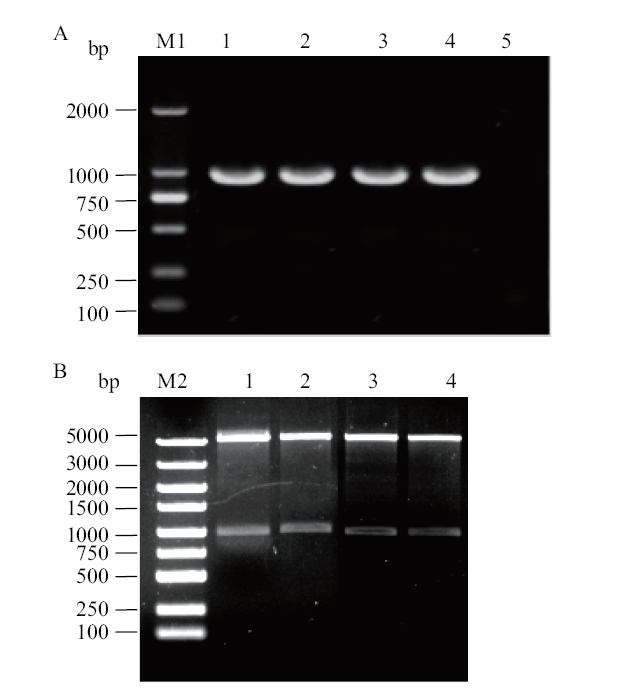
Fig. 1 PCR amplification of GapC gene and double digestion of recombinant plasmid A:PCR amplification of GapC gene;M1:DL 2000 DNA Marker;1-4: amplification of GapC gene,5: negative control. B:Double digestion of recombinant plasmid of GapC gene; M2:DL 5000 DNA Marker;1-4: double digestion of pET-28a-GapC
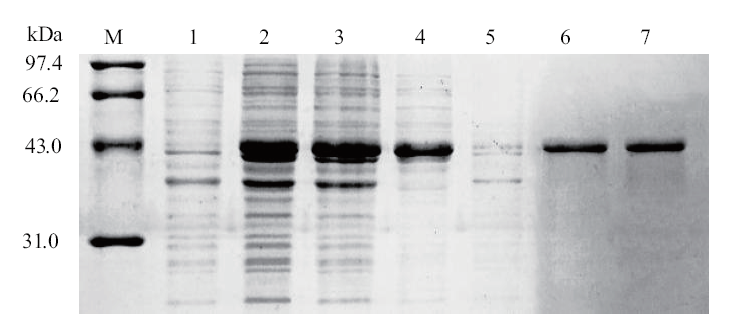
Fig. 2 Induction and purified result of recombinant GapC M: Protein marker; 1:uninduced by GapC; 2: after induced by GapC; 3: supernatant; 4: precipitation; 5: penetration; 6-7: elution
| 二级结构 Secondary structure | 占总氨基酸比例 Percentage in total amino acids/% |
|---|---|
| Hh: α-螺旋 | 31.07 |
| Ee: β-片层 | 25.74 |
| Tt: β-转角 | 9.47 |
| Cc: 无规则卷曲 | 33.73 |
Table 1 Secondary structure prediction of recombinant GapC protein
| 二级结构 Secondary structure | 占总氨基酸比例 Percentage in total amino acids/% |
|---|---|
| Hh: α-螺旋 | 31.07 |
| Ee: β-片层 | 25.74 |
| Tt: β-转角 | 9.47 |
| Cc: 无规则卷曲 | 33.73 |
| 多肽 Polypeptide | 多肽位置 Polypeptide position | 多肽序列 Polypeptide sequence |
|---|---|---|
| PL1 | 66-90 | RVNGKEVKSFSEPDASKLPWKDLNI |
| PL2 | 95-115 | ECTGFYTDKDKAQAHIEAGAK |
| PL4 | 181-208 | TGDQNTQDAPHRKGDKRRARAA AENIIP |
| PL5 | 221-236 | IPEIDGKLDGGAQRVP |
| PL6 | 247-268 | VVLEKQDVTVEQVNEAMKNASN |
| PL7 | 264-286 | KNASNESFGYTEDEIVSSDVVGM |
| PC8 | 297-308 | TRVMSVGDRQLV |
Table 2 Prediction of antigen epitope for recombinant GapC protein
| 多肽 Polypeptide | 多肽位置 Polypeptide position | 多肽序列 Polypeptide sequence |
|---|---|---|
| PL1 | 66-90 | RVNGKEVKSFSEPDASKLPWKDLNI |
| PL2 | 95-115 | ECTGFYTDKDKAQAHIEAGAK |
| PL4 | 181-208 | TGDQNTQDAPHRKGDKRRARAA AENIIP |
| PL5 | 221-236 | IPEIDGKLDGGAQRVP |
| PL6 | 247-268 | VVLEKQDVTVEQVNEAMKNASN |
| PL7 | 264-286 | KNASNESFGYTEDEIVSSDVVGM |
| PC8 | 297-308 | TRVMSVGDRQLV |
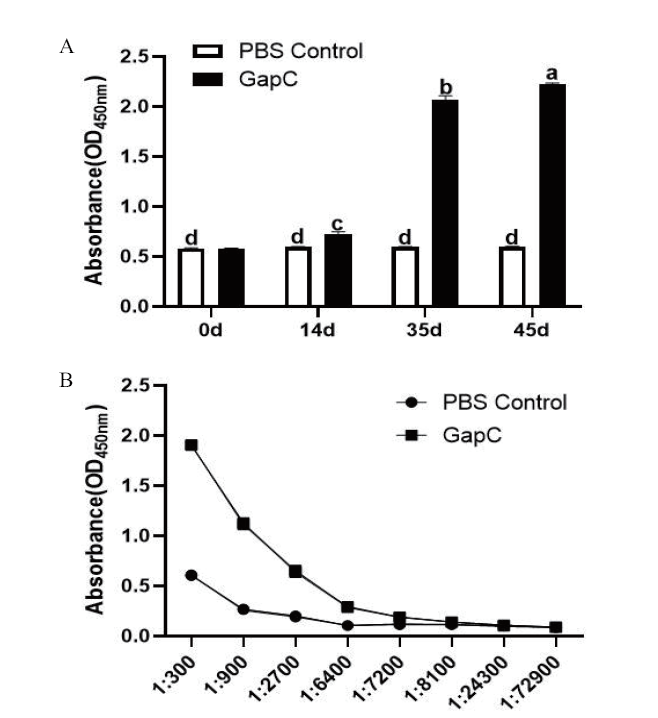
Fig. 7 Immunized rabbit serum antibody levels and titer detection results of recombinant GapC A:Immunized rabbit serum antibody levels detection results. B:Immunized rabbit serum antibody levels detection results, and different letters indicate significant differences between groups(P<0.05), the same below
| [7] |
Bradley AJ, Breen JE, Payne B, et al. An investigation of the efficacy of a polyvalent mastitis vaccine using different vaccination regimens under field conditions in the United Kingdom[J]. J Dairy Sci, 2015, 98(3): 1706-1720.
doi: 10.3168/jds.2014-8332 pmid: 25529419 |
| [8] |
Cheng BL, Nielsen TB, Pantapalangkoor P, et al. Evaluation of serotypes 5 and 8 capsular polysaccharides in protection against Staphylococcus aureus in murine models of infection[J]. Hum Vaccin Immunother, 2017, 13(7): 1609-1614.
doi: 10.1080/21645515.2017.1304334 URL |
| [9] |
Misra N, Wines TF, Knopp CL, et al. Immunogenicity of a Staphylococcus aureus-cholera toxin A2/B vaccine for bovine mastitis[J]. Vaccine, 2018, 36(24): 3513-3521.
doi: S0264-410X(18)30564-4 pmid: 29739718 |
| [10] | 程旭, 杨雨睛, 吴赛男, 等. 金黄色葡萄球菌SarA、IcaA及其融合基因的DNA疫苗构建及在小鼠免疫应答中的初步研究[J]. 中国生物工程杂志, 2020, 40(7): 41-50. |
| Cheng X, Yang YQ, Wu SN, et al. Construction of DNA vaccines of Staphylococcus aureus sar A, IcaA and their fusion genes and preliminary study in mouse immune response[J]. China Biotechnol, 2020, 40(7): 41-50. | |
| [11] |
Ma ZC, Yin XY, Wu P, et al. The recombinant expression proteins FnBP and ClfA from Staphylococcus aureus in addition to GapC and sip from Streptococcus agalactiae can protect BALB/c mice from bacterial infection[J]. Front Vet Sci, 2021, 8: 666098.
doi: 10.3389/fvets.2021.666098 URL |
| [12] |
Hajighahramani N, Nezafat N, Eslami M, et al. Immunoinformatics analysis and in silico designing of a novel multi-epitope peptide vaccine against Staphylococcus aureus[J]. Infect Genet Evol, 2017, 48: 83-94.
doi: S1567-1348(16)30528-7 pmid: 27989662 |
| [13] |
Zhao Z, Sun HQ, Wei SS, et al. Multiple B-cell epitope vaccine induces a Staphylococcus enterotoxin B-specific IgG1 protective response against MRSA infection[J]. Sci Rep, 2015, 5: 12371.
doi: 10.1038/srep12371 |
| [14] |
Yang SY, Li WY, Fan ZW, et al. Identification of CD4+ T cell epitopes on glyceraldehyde-3-phosphate dehydrogenase-C of Staphylococcus aureus in Babl/c mice[J]. Microb Pathog, 2020, 144: 104167.
doi: 10.1016/j.micpath.2020.104167 URL |
| [15] | 王汉青, 张雪静, 张欢, 等. 乳房链球菌GapC蛋白的表达及其B细胞抗原表位的预测与鉴定[J]. 生物工程学报, 2022, 38(1): 148-159. |
| Wang HQ, Zhang XJ, Zhang H, et al. Prokaryotic expression of the GapC protein of Streptococcus uberis and prediction, identification of its B-cell epitopes[J]. Chin J Biotechnol, 2022, 38(1): 148-159. | |
| [16] |
Middleton JR. Staphylococcus aureus antigens and challenges in vaccine development[J]. Expert Rev Vaccines, 2008, 7(6): 805-815.
doi: 10.1586/14760584.7.6.805 pmid: 18665778 |
| [17] | 赵达, 王鹤, 梁宏儒, 等. 金黄色葡萄球菌GapC蛋白与鼠伤寒沙门菌鞭毛蛋白的融合表达及其活性[J]. 中国生物制品学杂志, 2014, 27(2): 168-171, 176. |
| Zhao D, Wang H, Liang HR, et al. Fusion expression of GapC protein of Staphylococcus aureus and flagellin of Salmonella typhimurium and activity of expressed product[J]. Chin J Biol, 2014, 27(2): 168-171, 176. | |
| [18] |
Wang MY, Wei YH, Yu W, et al. Identification of a conserved linear B-cell epitope in the Staphylococcus aureus GapC protein[J]. Microb Pathog, 2018, 118: 39-47.
doi: 10.1016/j.micpath.2018.03.007 URL |
| [19] |
Landin H, Mörk MJ, Larsson M, et al. Vaccination against Staphylococcus aureus mastitis in two Swedish dairy herds[J]. Acta Vet Scand, 2015, 57: 81.
doi: 10.1186/s13028-015-0171-6 pmid: 26608421 |
| [20] |
Vinod N, Oh S, Park HJ, et al. Generation of a novel Staphylococcus aureus ghost vaccine and examination of its immunogenicity against virulent challenge in rats[J]. Infect Immun, 2015, 83(7): 2957-2965.
doi: 10.1128/IAI.00009-15 URL |
| [21] | 杨汐静, 陈晓婷, 刘道龙, 等. 金黄色葡萄球菌FnbpA免疫优势片段的构建及免疫保护性研究[J]. 重庆医学, 2020, 49(5): 813-818. |
| Yang XJ, Chen XT, Liu DL, et al. Study on construction of Staphylococcus aureus FnbpA immunodominant fragment and its protective rate[J]. Chongqing Med, 2020, 49(5): 813-818. | |
| [22] |
Zhang LM, Zhang H, Fan ZY, et al. Identification of a conserved linear B-cell epitope of Streptococcus dysgalactiae GapC protein by screening phage-displayed random peptide library[J]. PLoS One, 2015, 10(6): e0131221.
doi: 10.1371/journal.pone.0131221 URL |
| [23] |
Yao D, Zhang H, Wang XT, et al. Identification and characterization of CD4+ T-cell epitopes on GapC protein of Streptococcus dysgalactiae[J]. Microb Pathog, 2016, 91: 46-53.
doi: 10.1016/j.micpath.2015.11.025 URL |
| [24] |
Foster TJ, Geoghegan JA, Ganesh VK, et al. Adhesion, invasion and evasion: the many functions of the surface proteins of Staphylococcus aureus[J]. Nat Rev Microbiol, 2014, 12(1): 49-62.
doi: 10.1038/nrmicro3161 |
| [25] |
Kerro-Dego O, Prysliak T, Perez-Casal J, et al. Role of GapC in the pathogenesis of Staphylococcus aureus[J]. Vet Microbiol, 2012, 156(3/4): 443-447.
doi: 10.1016/j.vetmic.2011.11.018 URL |
| [26] |
Perez-Casal J, Prysliak T, Kerro-Dego O, et al. Immune responses to a Staphylococcus aureus GapC/B chimera and its potential use as a component of a vaccine for S. aureus mastitis[J]. Vet Immunol Immunopathol, 2006, 109(1-2): 85-97.
doi: 10.1016/j.vetimm.2005.07.024 URL |
| [27] | 朱洪伟, 朱战波, 崔玉东, 等. 金黄色葡萄球菌重组GapC蛋白的GAPDH活性及免疫原性分析[J]. 生物工程学报, 2008, 24(05):754-759. |
| Zhu HW, Zhu ZB, Cui YD, et al. GAPDH Activity and Immunogenicity of Staphylococcus aureus Recombinant GapC Protein[J]. Chin J Biotechn, 2008, 24(05):754-759. | |
| [28] | Closter JM, Bjoern P, Morten N, et al. BepiPred-2.0: improving sequence-based B-cell epitope prediction using conformational epitopes[J]. Nucleic Acids Res, 2017, 45(01):24-29. |
| [29] |
Saha S, Raghava GPS. Prediction of continuous B-cell epitopes in an antigen using recurrent neural network[J]. Proteins, 2006, 65(1): 40-48.
doi: 10.1002/prot.21078 URL |
| [30] | 孔智翔, 刘祥, 殴莎莎, 等. 奶牛乳房炎链球菌GapC蛋白的生物信息学分析与重组表位疫苗设计[J]. 江苏农业学报, 2016, 32(4): 824-831. |
| Kong ZX, Liu X, Ou SS, et al. Bioinformatics analysis of cow mastitis Streptococcus GapC protein and designing of recombinant epitope vaccine[J]. Jiangsu J Agric Sci, 2016, 32(4): 824-831. | |
| [1] | Lucia M, Rahayu S, Haerah D, et al. Detection of Staphylococcus aureus and Streptococcus agalactiae: subclinical mastitis causes in dairy cow and dairy buffalo(Bubalus bubalis)[J]. Amer J Biomed Res, 2017, 5(1):8-13. |
| [2] | 龙木措. 奶牛乳房炎的病因、治疗及预防[J]. 中国动物保健, 2021, 23(12): 21-22. |
| Long MC. Etiology, treatment and prevention of mastitis in dairy cows[J]. China Animal Heal, 2021, 23(12): 21-22. | |
| [3] |
Li LP, Zhao X. Characterization of the resistance class 1 integrons in Staphylococcus aureus isolates from milk of lactating dairy cattle in Northwestern China[J]. BMC Vet Res, 2018, 14(1): 59.
doi: 10.1186/s12917-018-1376-5 |
| [4] |
Awad A, Ramadan H, Nasr S, et al. Genetic characterization, antimicrobial resistance patterns and virulence determinants of Staphylococcus aureus isolated form bovine mastitis[J]. Pak J Biol Sci, 2017, 20(6): 298-305.
doi: 10.3923/pjbs.2017.298.305 URL |
| [5] |
Ronco T, Klaas IC, Stegger M, et al. Genomic investigation of Staphylococcus aureus isolates from bulk tank milk and dairy cows with clinical mastitis[J]. Vet Microbiol, 2018, 215: 35-42.
doi: 10.1016/j.vetmic.2018.01.003 URL |
| [6] |
Nagasawa Y, Kiku Y, Sugawara K, et al. Exfoliation rate of mammary epithelial cells in milk on bovine mastitis caused by Staphylococcus aureus is associated with bacterial load[J]. Anim Sci J, 2018, 89(1): 259-266.
doi: 10.1111/asj.12886 pmid: 28891152 |
| [31] | 张丽萌, 周雪, 魏玉华, 等. 停乳链球菌GapC1-150aa蛋白单克隆抗体的制备及其线性表位鉴定[J]. 中国预防兽医学报, 2015, 37(11): 875-880. |
| Zhang LM, Zhou X, Wei YH, et al. Preparation of monoclonal antibodies against Streptococcus dysgalactiae GapC1-150aa and identification of a linear epitope to the MAbs[J]. Chin J Prev Vet Med, 2015, 37(11): 875-880. | |
| [32] |
Ebrahimi SM, Tebianian M, Aghaiypour K, et al. Prokaryotic expression and characterization of avian influenza A virus M2 gene as a candidate for universal recombinant vaccine against influenza A subtypes; specially H5N1 and H9N2[J]. Mol Biol Rep, 2010, 37(6): 2909-2914.
doi: 10.1007/s11033-009-9851-5 pmid: 19809890 |
| [33] | 马骏, 王然, 刘硕, 等. 基于金黄色葡萄球菌FnBPA和停乳链球菌GapC的多表位疫苗免疫原性研究[J]. 黑龙江八一农垦大学学报, 2021, 33(1): 60-67, 114. |
| Ma J, Wang R, Liu S, et al. Immunogenicity of multi-epitope vaccines based on S. aureus FnBPA and S. dysgalactiae GapC[J]. J Heilongjiang Bayi Agric Univ, 2021, 33(1): 60-67, 114. |
| [1] | LIN Hong-yan, GUO Xiao-rui, LIU Di, LI Hui, LU Hai. Molecular Mechanism of Transcriptional Factor AtbHLH68 in Regulating Cell Wall Development by Transcriptome Analysis [J]. Biotechnology Bulletin, 2023, 39(9): 105-116. |
| [2] | YANG Zhi-xiao, HOU Qian, LIU Guo-quan, LU Zhi-gang, CAO Yi, GOU Jian-yu, WANG Yi, LIN Ying-chao. Responses of Rubisco and Rubisco Activase in Different Resistant Tobacco Strains to Brown Spot Stress [J]. Biotechnology Bulletin, 2023, 39(9): 202-212. |
| [3] | CHEN Zhong-yuan, WANG Yu-hong, DAI Wei-jun, ZHANG Yan-min, YE Qian, LIU Xu-ping, TAN Wen-Song, ZHAO Liang. Mechanism Investigation of Ferric Ammonium Citrate on Transfection for Suspended HEK293 Cells [J]. Biotechnology Bulletin, 2023, 39(9): 311-318. |
| [4] | WANG Jia-rui, SUN Pei-yuan, KE Jin, RAN Bin, LI Hong-you. Cloning and Expression Analyses of C-glycosyltransferase Gene FtUGT143 in Fagopyrum tataricum [J]. Biotechnology Bulletin, 2023, 39(8): 204-212. |
| [5] | LI Bo, LIU He-xia, CHEN Yu-ling, ZHOU Xing-wen, ZHU Yu-lin. Cloning, Subcellular Localization and Expression Analysis of CnbHLH79 Transcription Factor from Camellia nitidissima [J]. Biotechnology Bulletin, 2023, 39(8): 241-250. |
| [6] | WANG Shuai, FENG Yu-mei, BAI Miao, DU Wei-jun, YUE Ai-qin. Functional Analysis of Soybean Gene GmHMGR Responding to Exogenous Hormones and Abiotic Stresses [J]. Biotechnology Bulletin, 2023, 39(7): 131-142. |
| [7] | SUN Ming-hui, WU Qiong, LIU Dan-dan, JIAO Xiao-yu, WANG Wen-jie. Cloning and Expression Analysis of CsTMFs Gene in Tea Plant [J]. Biotechnology Bulletin, 2023, 39(7): 151-159. |
| [8] | MEI Huan, LI Yue, LIU Ke-meng, LIU Ji-hua. Study on the Biosynthesis of l-SLR by Efficient Prokaryotic Expression of Berberine Bridge Enzyme [J]. Biotechnology Bulletin, 2023, 39(7): 277-287. |
| [9] | ZHAO Xue-ting, GAO Li-yan, WANG Jun-gang, SHEN Qing-qing, ZHANG Shu-zhen, LI Fu-sheng. Cloning and Expression of AP2/ERF Transcription Factor Gene ShERF3 in Sugarcane and Subcellular Localization of Its Encoded Protein [J]. Biotechnology Bulletin, 2023, 39(6): 208-216. |
| [10] | ZHANG Lu-yang, HAN Wen-long, XU Xiao-wen, YAO Jian, LI Fang-fang, TIAN Xiao-yuan, ZHANG Zhi-qiang. Identification and Expression Analysis of the Tobacco TCP Gene Family [J]. Biotechnology Bulletin, 2023, 39(6): 248-258. |
| [11] | QIAN Bang, LIU Zhen-dong, ZHAO Yin, LI Jing, PRAJAPATI Meera, LI Yan-min, SUN Yue-feng, DOU Yong-xi. Establishment of Chemiluminescence Immunoassay for the Detection of Peste des Petits Ruminants Virus H Protein Antibodies [J]. Biotechnology Bulletin, 2023, 39(5): 120-129. |
| [12] | LI Zhi-qi, YUAN Yue, MIAO Rong-qing, PANG Qiu-ying, ZHANG Ai-qin. Melatonin Contents in Eutrema salsugineum and Arabidopsis thaliana Under Salt Stress, and Expression Pattern Analysis of Synthesis Related Genes [J]. Biotechnology Bulletin, 2023, 39(5): 142-151. |
| [13] | LI Jing-rui, WANG Yu-bo, XIE Zi-wei, LI Chang, WU Xiao-lei, GONG Bin-bin, GAO Hong-bo. Identification and Expression Analysis of PIN Gene Family in Melon Under High Temperature Stress [J]. Biotechnology Bulletin, 2023, 39(5): 192-204. |
| [14] | LIU Kui, LI Xing-fen, YANG Pei-xin, ZHONG Zhao-chen, CAO Yi-bo, ZHANG Ling-yun. Functional Study and Validation of Transcriptional Coactivator PwMBF1c in Picea wilsonii [J]. Biotechnology Bulletin, 2023, 39(5): 205-216. |
| [15] | JIANG Qing-chun, DU Jie, WANG Jia-cheng, YU Zhi-he, WANG Yun, LIU Zhong-yu. Expression and Function Analysis of Transcription Factor PcMYB2 from Polygonum cuspidatum [J]. Biotechnology Bulletin, 2023, 39(5): 217-223. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||