








Biotechnology Bulletin ›› 2023, Vol. 39 ›› Issue (6): 259-273.doi: 10.13560/j.cnki.biotech.bull.1985.2022-1316
Previous Articles Next Articles
YANG Yang1( ), ZHU Jin-cheng1, LOU Hui1, HAN Ze-gang2(
), ZHU Jin-cheng1, LOU Hui1, HAN Ze-gang2( ), ZHANG Wei1(
), ZHANG Wei1( )
)
Received:2022-10-26
Online:2023-06-26
Published:2023-07-07
Contact:
HAN Ze-gang, ZHANG Wei
E-mail:1977277872@qq.com;zeganghan@zju.edu.cn;zhw_agr@shzu.edu.cn
YANG Yang, ZHU Jin-cheng, LOU Hui, HAN Ze-gang, ZHANG Wei. Transcriptome Analysis of Interaction Between Gossypium barbadense and Fusarium oxysporum f. sp. vasinfectum[J]. Biotechnology Bulletin, 2023, 39(6): 259-273.
| 基因名称Gene name | 上游引物 Forward primer(5'-3') | 下游引物 Reverse primer(5'-3') |
|---|---|---|
| Gb2763 | TTCCACAGTTCCAGGGTA | GATGATCCAAGGCTTTCT |
| Gb1474 | AAACAAGGTAAGGAGGCT | GTTGGGTAAATAATAGGC |
| Gb7593 | CAACTTGGCATCTTATCC | CTTACATTGAGCAGCAAC |
| Gb8190 | CATGGAGCAAAGGTTTAA | TGTTGGATTCAGGGAGAT |
| Gb8191 | CATGGAGCAAAGGTTTAA | TGTTGGATTCAGGGAGAT |
| Gb9261 | TTTGCCAAAGAGCGTAGA | TTAAAGAATGCGGTGTCG |
| Gb9263 | TTTCGGCAAGTCTATGAT | TCTTTCTGGTCGTGGATG |
| FOTG10073 | GATGCCTGCCGTGGTGGAT | GGGCGAAGAAACAGTGTAAAGC |
| FOTG05109 | TATCGCCCAGATGTTCAA | TACCACGGTGCTGTTCCA |
| FOTG15482 | TTTCTTCATCGCCCTGTA | ACATTGCCGACTTGGATT |
| FOTG11725 | TTGTTGGTTTGCCCTTGT | CGAGCGGTCTGAGTGTAG |
| FOTG17884 | GCAATCTCAGCCTCAAGTT | GACAAGCGTTCATCCATAA |
| FOTG17663 | GCCTTCTCGGATCTTCTA | GCAATGCTGGGATGGTAT |
| GhUBQ7 | GAAGACCTACACCAAGCCCAA | CGGACTCTACTCAATCCCCACC |
Table 1 Primers for RT-qPCR
| 基因名称Gene name | 上游引物 Forward primer(5'-3') | 下游引物 Reverse primer(5'-3') |
|---|---|---|
| Gb2763 | TTCCACAGTTCCAGGGTA | GATGATCCAAGGCTTTCT |
| Gb1474 | AAACAAGGTAAGGAGGCT | GTTGGGTAAATAATAGGC |
| Gb7593 | CAACTTGGCATCTTATCC | CTTACATTGAGCAGCAAC |
| Gb8190 | CATGGAGCAAAGGTTTAA | TGTTGGATTCAGGGAGAT |
| Gb8191 | CATGGAGCAAAGGTTTAA | TGTTGGATTCAGGGAGAT |
| Gb9261 | TTTGCCAAAGAGCGTAGA | TTAAAGAATGCGGTGTCG |
| Gb9263 | TTTCGGCAAGTCTATGAT | TCTTTCTGGTCGTGGATG |
| FOTG10073 | GATGCCTGCCGTGGTGGAT | GGGCGAAGAAACAGTGTAAAGC |
| FOTG05109 | TATCGCCCAGATGTTCAA | TACCACGGTGCTGTTCCA |
| FOTG15482 | TTTCTTCATCGCCCTGTA | ACATTGCCGACTTGGATT |
| FOTG11725 | TTGTTGGTTTGCCCTTGT | CGAGCGGTCTGAGTGTAG |
| FOTG17884 | GCAATCTCAGCCTCAAGTT | GACAAGCGTTCATCCATAA |
| FOTG17663 | GCCTTCTCGGATCTTCTA | GCAATGCTGGGATGGTAT |
| GhUBQ7 | GAAGACCTACACCAAGCCCAA | CGGACTCTACTCAATCCCCACC |
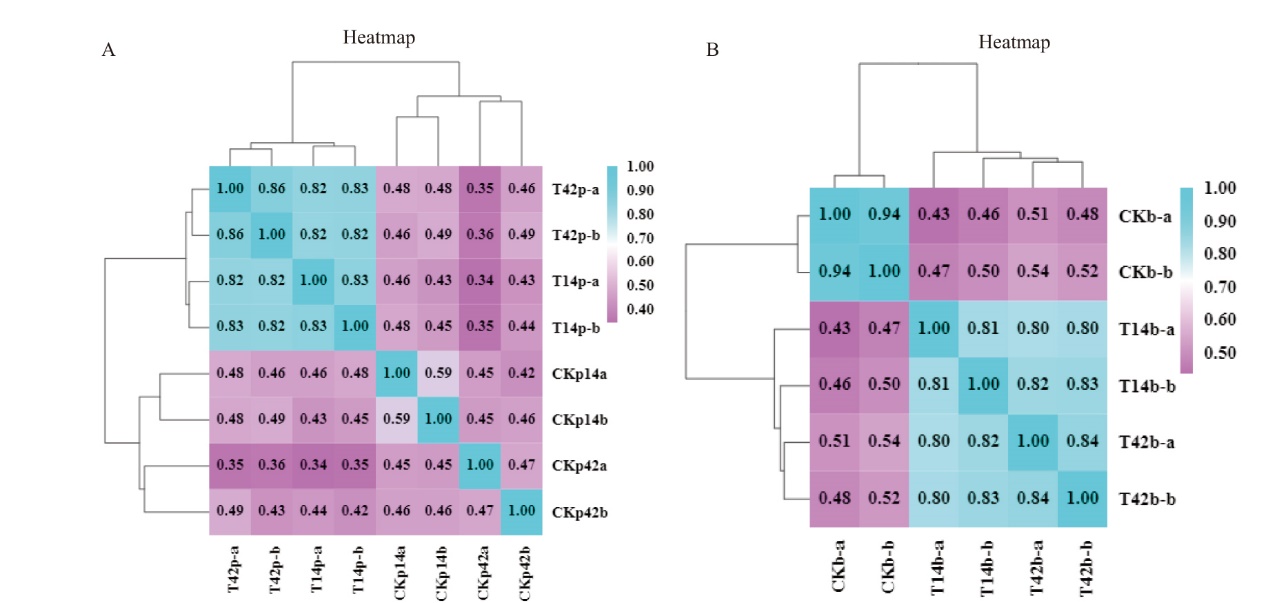
Fig. 1 Correlation analysis among samples A: Correlation analysis of four samples from cotton root tissues. T14p indicates the treatment sample from Xinhai 14, T42p indicates the treatment sample from Xinhai 42, CKp14 indicates the control sample from Xinhai 14 and CKp42 indicates the control sample from Xinhai 42. B: Correlation analysis of three samples from Fov. CKb refers to the control sample of Fov, T14b to the Fov sample obtained from Xinhai 14, T42b to the Fov sample obtained from Xinhai 42. a and b are the duplications
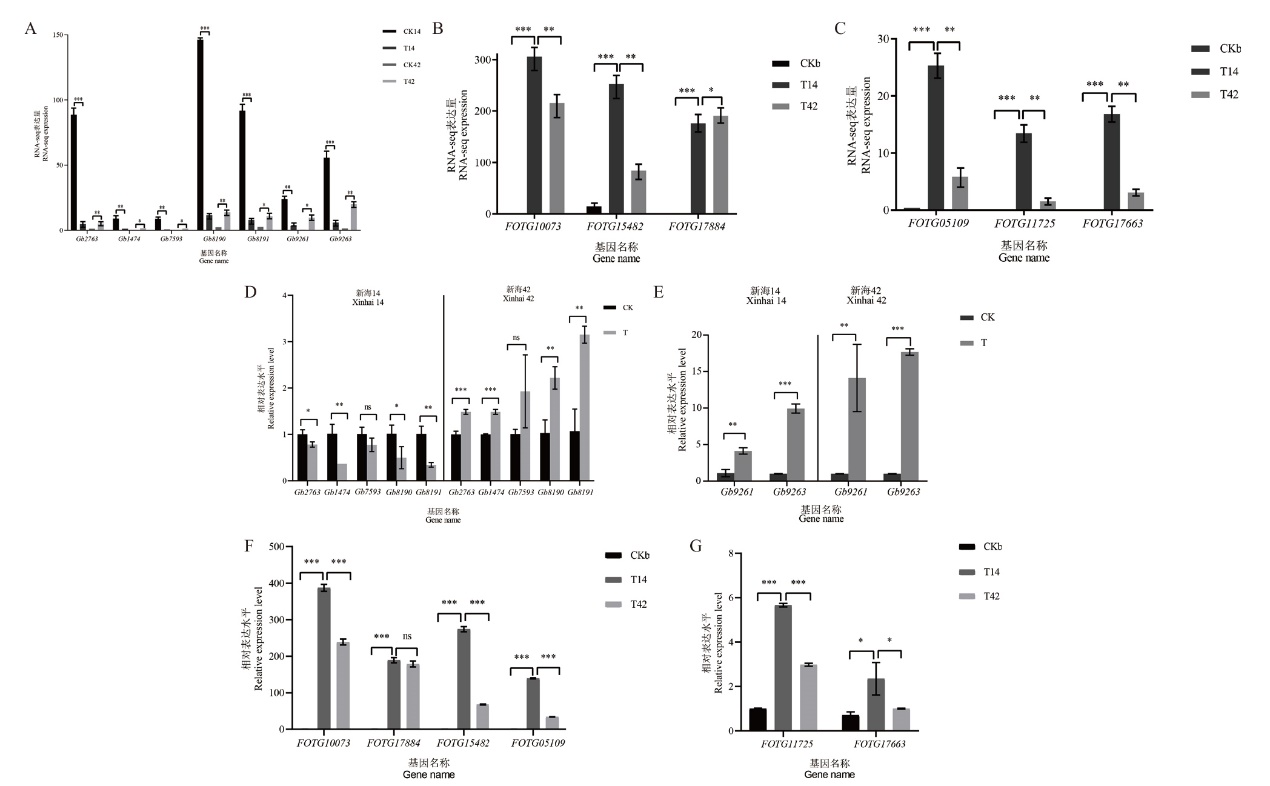
Fig. 2 Validation of RNA-seq data by RT-qPCR A-C : RNA-seq expression. D and E : RT-qPCR verification of 7 genes in cotton. F and G : RT-qPCR verification of 6 genes in Fusarium oxysporum. CK: Control group. T: Treatment group. CKb: Bacteria control group. T14: Xinhai 14 treatment group.T42: Xinhai 42 treatment group. * indicates difference(P<0.05), ** indicates significant difference(P<0.01), and *** indicates extremely significant difference(P<0.001)
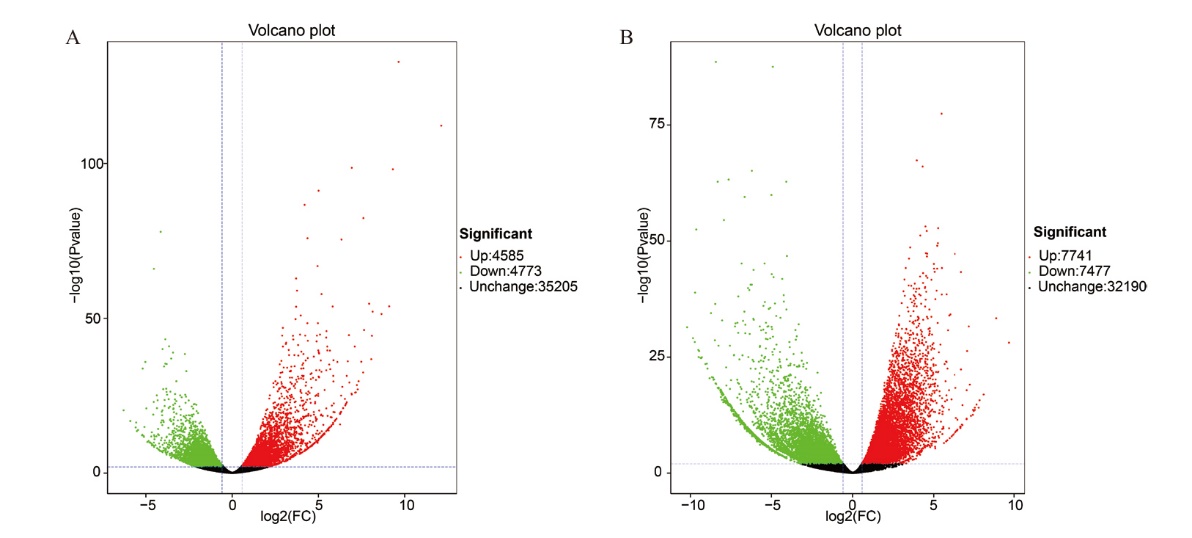
Fig. 3 Volcano map of differentially expressed genes in the cotton of different treatments Green is down-regulated gene and red is up- regulated gene. The same below

Fig. 5 GO analysis of differentially expressed genes in Xinhai 14 after inoculation A-C: GO analysis of up-regulated genes. D-F: GO analysis of down-regulated genes. The abscissa is number of genes enriched for each term, and the ordinate is the term of GO level
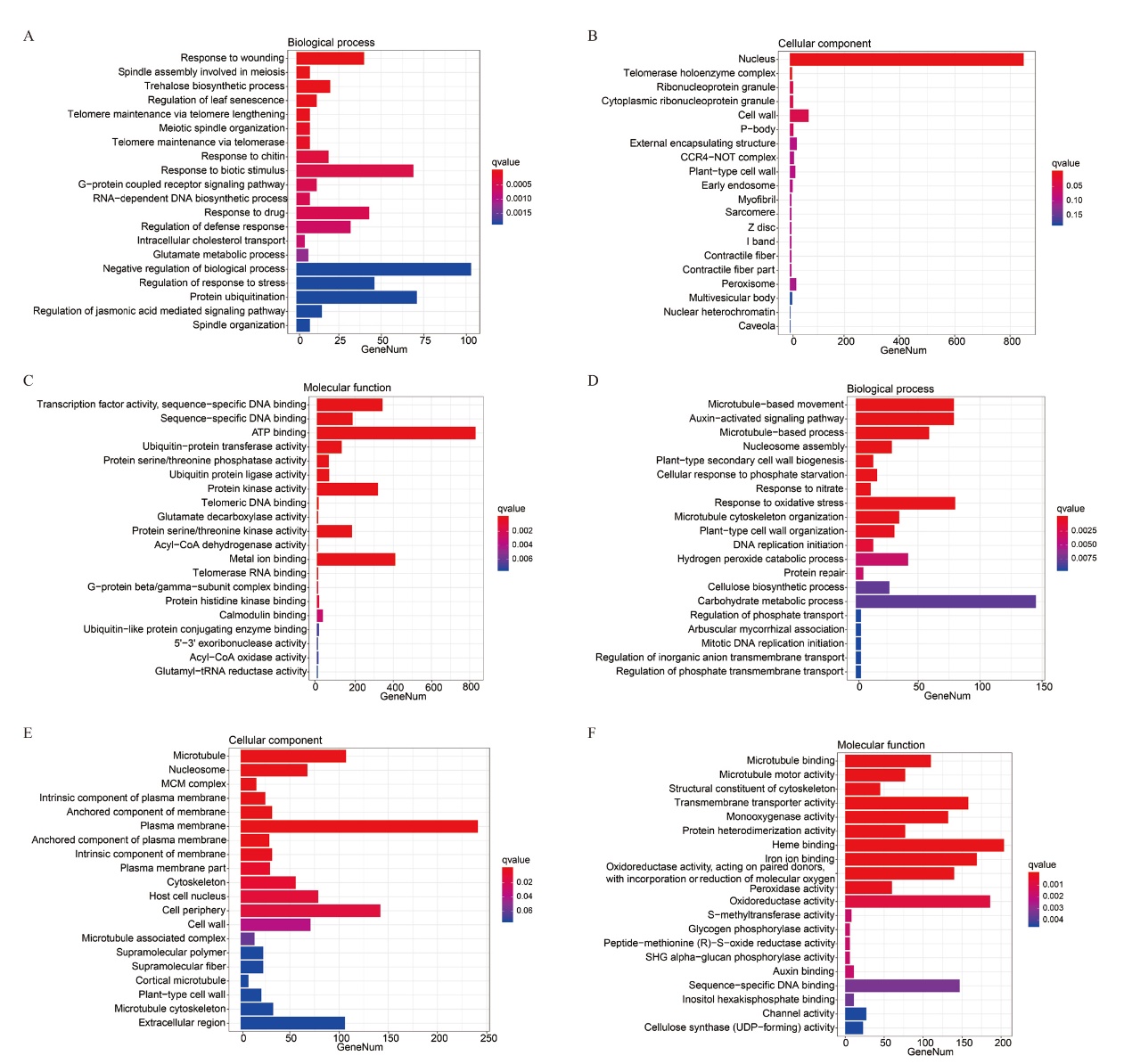
Fig. 6 GO analysis of differentially expressed genes in Xinhai 42 after inoculation A-C: GO analysis of up-regulated genes. D-F: GO analysis of down-regulated genes. The abscissa is number of genes enriched for each term,the ordinate is the term of GO level
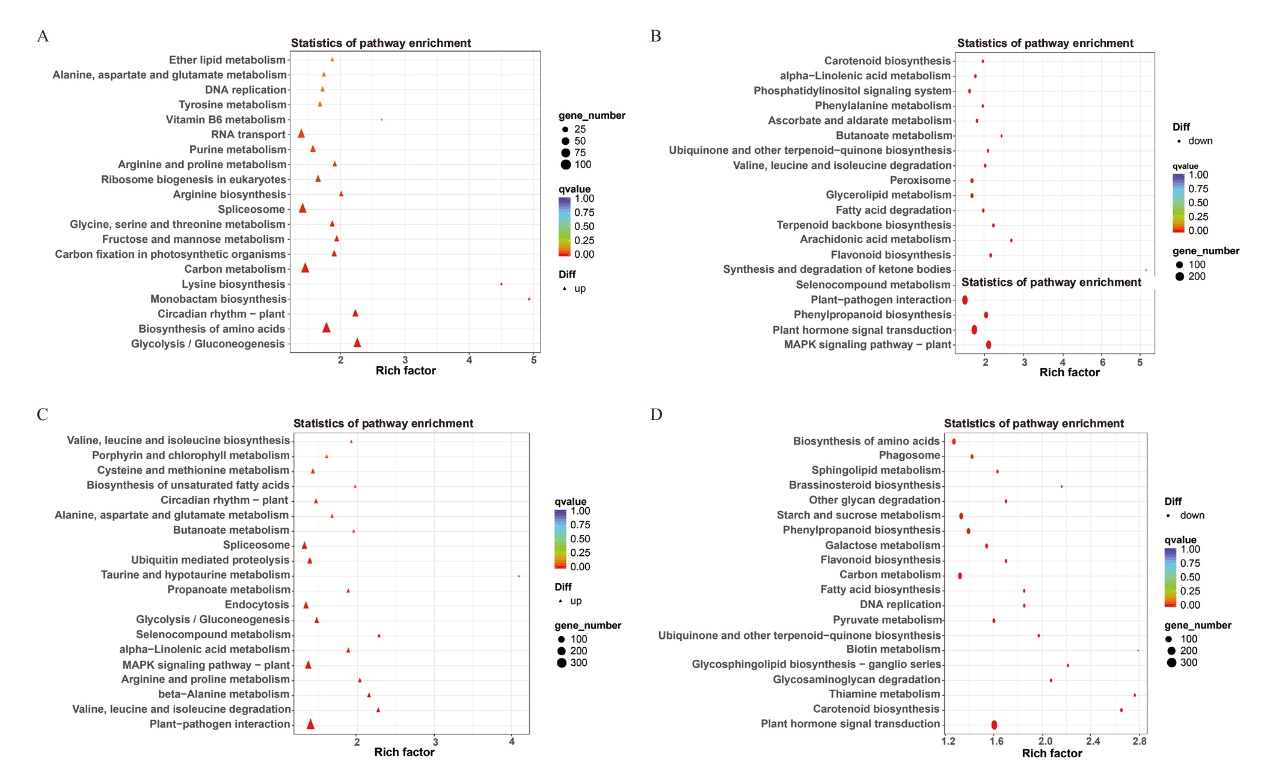
Fig. 7 KEGG analysis of differentially expressed genes in two cotton cultivars after inoculation A: KEGG analysis of up-regulated genes in Xinhai 14. B: KEGG analysis of down-regulated genes in Xinhai 14. C: KEGG analysis of up-regulated genes in Xinhai 42. D: KEGG analysis of down-regulated genes in Xinhai 42

Fig. 8 GO analysis of differentially expressed genes of Fov after infecting Xinhai 14 A-C: GO analysis of up-regulated genes of Fov in Xinhai 14. D-F: GO analysis of down-regulated genes of Fov in Xinhai 14. The abscissa is number of genes enriched for each term, the ordinate is the term of GO level
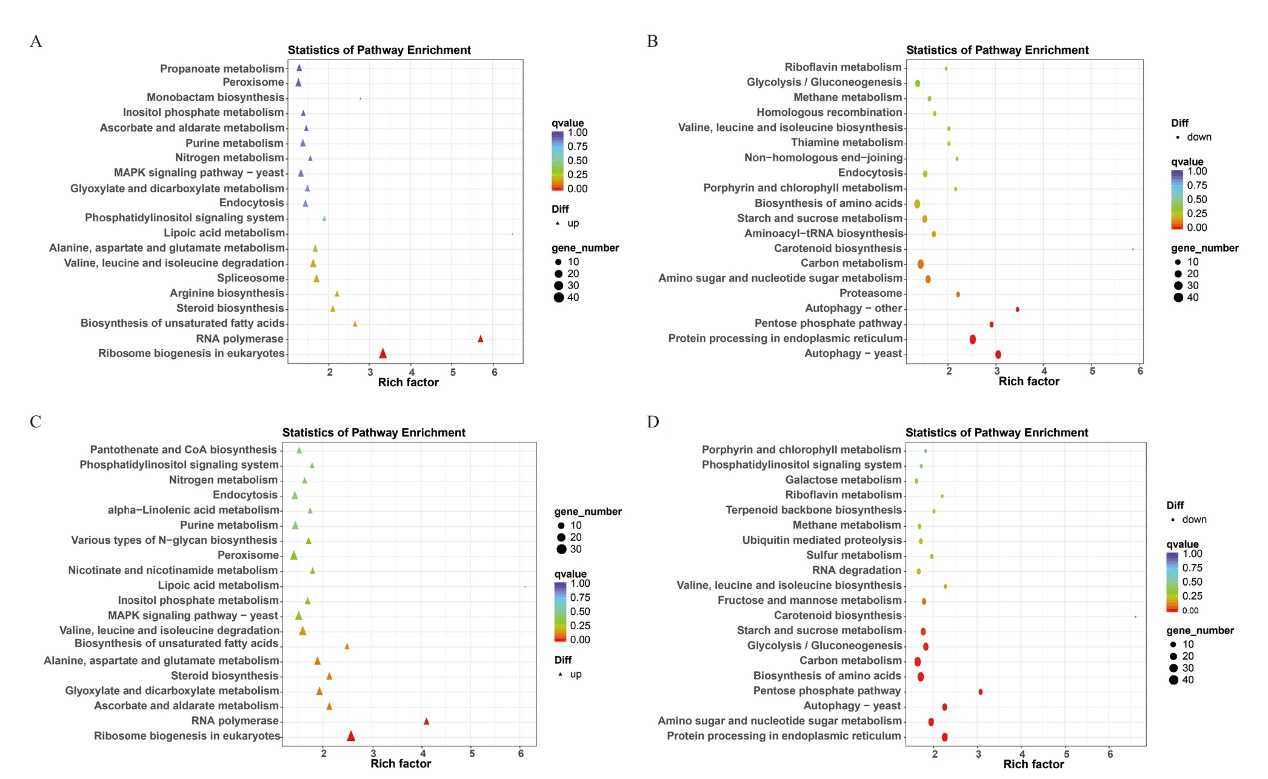
Fig. 9 KEGG analysis of differentially expressed genes of Fov after infecting Xinhai 14 and Xinhai 42 A: KEGG analysis of up-regulated genes of Fov(Fusarium oxysporum f. sp. Vasinfectum)in Xinhai 14. B: KEGG analysis of down-regulated genes of Fov in Xinhai 14. C: KEGG analysis of up-regulated genes of Fov in Xinhai 42. D: KEGG analysis of down-regulated genes of Fov in Xinhai 42

Fig. 10 GO analysis of differentially expressed genes of Fov after infecting Xinhai 42 A-C: GO analysis of up-regulated genes of Fov in Xinhai 42. D-F: GO analysis of down-regulated genes of Fov in Xinhai 42. The abscissa is number of genes enriched for each term, the ordinate is the term of GO level
| [16] | 雷忠华. 基于互作转录组分析的抗病香蕉品系南天蕉与尖孢镰刀菌互作机理研究[D]. 广州: 华南理工大学, 2018. |
| Lei ZH. Dual RNA-sequencing unveils host-pathogen interactions in resistant banana cultivars Nantianjiao upon Fusarium oxysporum race 4 challenge[D]. Guangzhou: South China University of Technology, 2018. | |
| [17] |
Ma ZY, Zhang Y, Wu LQ, et al. High-quality genome assembly and resequencing of modern cotton cultivars provide resources for crop improvement[J]. Nat Genet, 2021, 53(9): 1385-1391.
doi: 10.1038/s41588-021-00910-2 pmid: 34373642 |
| [18] |
刘戈辉, 韩泽刚, 孙士超, 等. 转GhB301基因棉花响应枯萎病菌侵染的转录组分析[J]. 核农学报, 2021, 35(12): 2733-2745.
doi: 10.11869/j.issn.100-8551.2021.12.2733 |
| Liu GH, Han ZG, Sun SC, et al. Transcriptome analysis of GhB301 transgenic cotton response to Fusarium wilt infection[J]. J Nucl Agric Sci, 2021, 35(12): 2733-2745. | |
| [19] |
Young MD, Wakefield MJ, Smyth GK, et al. Gene ontology analysis for RNA-seq: accounting for selection bias[J]. Genome Biol, 2010, 11(2): R14.
doi: 10.1186/gb-2010-11-2-r14 URL |
| [20] |
Kanehisa M, Goto S. KEGG: Kyoto encyclopedia of genes and genomes[J]. Nucleic Acids Res, 2000, 28(1): 27-30.
doi: 10.1093/nar/28.1.27 pmid: 10592173 |
| [21] |
Spoel SH, Dong XN. How do plants achieve immunity? Defence without specialized immune cells[J]. Nat Rev Immunol, 2012, 12(2): 89-100.
doi: 10.1038/nri3141 pmid: 22273771 |
| [22] |
Xiong DG, Wang YL, Ma J, et al. Deep mRNA sequencing reveals stage-specific transcriptome alterations during microsclerotia development in the smoke tree vascular wilt pathogen, Verticillium dahliae[J]. BMC Genomics, 2014, 15(1): 324.
doi: 10.1186/1471-2164-15-324 |
| [23] |
Berens ML, Berry HM, Mine A, et al. Evolution of hormone signaling networks in plant defense[J]. Annu Rev Phytopathol, 2017, 55: 401-425.
doi: 10.1146/annurev-phyto-080516-035544 pmid: 28645231 |
| [24] |
Bürger M, Chory J. Stressed out about hormones: how plants orchestrate immunity[J]. Cell Host Microbe, 2019, 26(2): 163-172.
doi: S1931-3128(19)30350-6 pmid: 31415749 |
| [1] |
Han ZG, Chen H, Cao YW, et al. Genomic insights into genetic improvement of upland cotton in the world's largest growing region[J]. Ind Crops Prod, 2022, 183: 114929.
doi: 10.1016/j.indcrop.2022.114929 URL |
| [2] | 朱永军, 张西英, 李金荣, 等. 海岛棉枯萎病抗性遗传规律及分子标记的初步筛选[J]. 新疆农业科学, 2010, 47(2): 268-273. |
| Zhu YJ, Zhang XY, Li JR, et al. Inheritance of resistance to Fusarium wilt and their molecular marker in Gossypium barbadense[J]. Xinjiang Agric Sci, 2010, 47(2): 268-273. | |
| [3] |
Michielse CB, Rep M. Pathogen profile update: Fusarium oxysporum[J]. Mol Plant Pathol, 2009, 10(3): 311-324.
doi: 10.1111/j.1364-3703.2009.00538.x pmid: 19400835 |
| [4] |
Back MA, Haydock PPJ, Jenkinson P. Disease complexes involving plant parasitic nematodes and soilborne pathogens[J]. Plant Pathol, 2002, 51(6): 683-697.
doi: 10.1046/j.1365-3059.2002.00785.x URL |
| [5] |
Wang C, Roberts PA. A Fusarium Wilt resistance gene in Gossypium barbadense and its effect on root-knot nematode-wilt disease complex[J]. Phytopathology, 2006, 96(7): 727-734.
doi: 10.1094/PHYTO-96-0727 pmid: 18943146 |
| [6] | 黄启秀. 海岛棉类黄酮代谢途径中与枯萎病抗性相关基因的克隆及功能验证[D]. 乌鲁木齐: 新疆农业大学, 2017. |
| Huang QX. Cloning and functional verification of genes related to Fusarium wilt resistance in flavonoid metabolism pathway of island cotton(Gossypium barbadense L.)[D]. Urumqi: Xinjiang Agricultural University, 2017. | |
| [7] | 赵曾强. 棉花GhWRKY44和GhWRKY22基因的克隆及功能初步分析[D]. 石河子: 石河子大学, 2015. |
| Zhao ZQ. Cloning and functional analysis of GhWRKY44 and GhWRKY22 gene in cotton[D]. Shihezi: Shihezi University, 2015. | |
| [8] | 张艳艳, 季苇芹, 杨玉文, 等. 转录组学技术及其在瓜菜作物上的应用[J]. 中国瓜菜, 2016, 29(12): 1-5, 13. |
| Zhang YY, Ji WQ, Yang YW, et al. The technology of transcriptome and its application in cucurbit and vegetable crops[J]. China Cucurbits Veg, 2016, 29(12): 1-5, 13. | |
| [9] |
Zhang WW, Zhang HC, Liu K, et al. Large-scale identification of Gossypium hirsutum genes associated with Verticillium dahliae by comparative transcriptomic and reverse genetics analysis[J]. PLoS One, 2017, 12(8): e0181609.
doi: 10.1371/journal.pone.0181609 URL |
| [10] |
Xu L, Zhu LF, Tu LL, et al. Lignin metabolism has a central role in the resistance of cotton to the wilt fungus Verticillium dahliae as revealed by RNA-Seq-dependent transcriptional analysis and histochemistry[J]. J Exp Bot, 2011, 62(15): 5607-5621.
doi: 10.1093/jxb/err245 pmid: 21862479 |
| [11] |
Jun Z, Zhang ZY, Gao YL, et al. Overexpression of GbRLK, a putative receptor-like kinase gene, improved cotton tolerance to Verticillium wilt[J]. Sci Rep, 2015, 5: 15048.
doi: 10.1038/srep15048 |
| [12] |
Zhang LS, Ni H, Du X, et al. The Verticillium-specific protein VdSCP7 localizes to the plant nucleus and modulates immunity to fungal infections[J]. New Phytol, 2017, 215(1): 368-381.
doi: 10.1111/nph.2017.215.issue-1 URL |
| [13] | 冯志迪. V型肌球蛋白在大丽轮枝菌与棉花互作过程中的机制研究[D]. 石河子: 石河子大学, 2017. |
| Feng ZD. Essential roles of the myosin V in plant cotton-verticillum interaction[D]. Shihezi: Shihezi University, 2017. | |
| [14] |
Soanes DM, Chakrabarti A, Paszkiewicz KH, et al. Genome-wide transcriptional profiling of appressorium development by the rice blast fungus Magnaporthe oryzae[J]. PLoS Pathog, 2012, 8(2): e1002514.
doi: 10.1371/journal.ppat.1002514 URL |
| [15] | 李成伟. 番茄与白粉菌互作的细胞学和转录组学分析[D]. 北京: 中国农业科学院, 2007. |
| Li CW. The cytological and transcriptomic investigations into interactions of tomato and powdery mildew[D]. Beijing: Chinese Academy of Agricultural Sciences, 2007. | |
| [25] |
Duressa D, Anchieta A, Chen DQ, et al. RNA-seq analyses of gene expression in the microsclerotia of Verticillium dahliae[J]. BMC Genomics, 2013, 14: 607.
doi: 10.1186/1471-2164-14-607 |
| [26] |
Zou Q, Luo S, Wu HT, et al. A GMC oxidoreductase GmcA is required for symbiotic nitrogen fixation in Rhizobium leguminosarum bv. viciae[J]. Front Microbiol, 2020, 11: 394.
doi: 10.3389/fmicb.2020.00394 URL |
| [27] | Arun KS, Vinay S, Jyoti S. A review on strain improvement of fungi for enhanced lipase production[J]. Journal of Biology and Nature, 2017, 6(3): 173-180. |
| [28] |
Kim YK, Wang YH, Liu ZM, et al. Identification of a hard surface contact-induced gene in Colletotrichum gloeosporioides conidia as a sterol glycosyl transferase, a novel fungal virulence factor[J]. Plant J, 2002, 30(2): 177-187.
doi: 10.1046/j.1365-313X.2002.01284.x URL |
| [29] |
Shen M, Zhao DK, Qiao Q, et al. Identification of glutathione S-transferase(GST)genes from a dark septate endophytic fungus(Exophiala pisciphila)and their expression patterns under varied metals stress[J]. PLoS One, 2015, 10(4): e0123418.
doi: 10.1371/journal.pone.0123418 URL |
| [1] | LOU Hui, ZHU Jin-cheng, YANG Yang, ZHANG Wei. Effects of Root Exudates in Resistant and Susceptible Varieties of Cotton on the Growths and Gene Expressions of Fusarium oxysporum [J]. Biotechnology Bulletin, 2023, 39(9): 156-167. |
| [2] | WU Yuan-ming, LIN Jia-yi, LIU Yu-xi, LI Dan-ting, ZHANG Zong-qiong, ZHENG Xiao-ming, PANG Hong-bo. Identification of Rice Plant Height-associated QTL Using BSA-seq and RNA-seq [J]. Biotechnology Bulletin, 2023, 39(8): 173-184. |
| [3] | FU Yu, JIA Rui-rui, HE He, WANG Liang-gui, YANG Xiu-lian. Growth Differences Among Grafted Seedlings with Two Rootstocks of Catalpa bungei and Comparative Analysis of Transcriptome [J]. Biotechnology Bulletin, 2023, 39(8): 251-261. |
| [4] | ZHAO Lin-yan, XU Wu-mei, WANG Hao-ji, WANG Kun-yan, WEI Fu-gang, YANG Shao-zhou, GUAN Hui-lin. Effects of Applying Biochar on the Rhizosphere Fungal Community and Survival Rate of Panax notoginseng Under Continuous Cropping [J]. Biotechnology Bulletin, 2023, 39(7): 219-227. |
| [5] | XIE Yang, XING Yu-meng, ZHOU Guo-yan, LIU Mei-yan, YIN Shan-shan, YAN Li-ying. Transcriptome Analysis of Diploid and Autotetraploid in Cucumber Fruit [J]. Biotechnology Bulletin, 2023, 39(3): 152-162. |
| [6] | SHEN Yun-xin, SHI Zhu-feng, ZHOU Xu-dong, LI Ming-gang, ZHANG Qing, FENG Lu-yao, CHEN Qi-bin, YANG Pei-wen. Isolation, Identification and Bio-activity of Three Bacillus Strains with Biocontrol Function [J]. Biotechnology Bulletin, 2023, 39(3): 267-277. |
| [7] | DENG Jia-hui, LEI Jian-feng, ZHAO Yi, LIU Min, HU Zi-yao, YOU Yang-zi, SHAO Wu-kui, LIU Jian-fei, LIU Xiao-dong. Construction of a New Mini Genome Editing System Based on Csy4 and MCP [J]. Biotechnology Bulletin, 2023, 39(10): 68-79. |
| [8] | ZHU Jin-cheng, YANG Yang, LOU Hui, ZHANG Wei. Regulation of Fusarium wilt Resistance in Cotton by Exogenous Melatonin [J]. Biotechnology Bulletin, 2023, 39(1): 243-252. |
| [9] | SUN Zhuo, WANG Yan, HAN Zhong-ming, WANG Yun-he, ZHAO Shu-jie, YANG Li-min. Isolation, Identification and Biocontrol Potential of Rhizospheric Fungus of Saposhnikovia divaricata [J]. Biotechnology Bulletin, 2023, 39(1): 264-273. |
| [10] | LI Ji-hong, JING Yu-ling, MA Gui-zhen, GUO Rong-jun, LI Shi-dong. Genome Construction of Achromobacter 77 and Its Characteristics on Chemotaxis and Antibiotic Resistance [J]. Biotechnology Bulletin, 2022, 38(9): 136-146. |
| [11] | LI Xiu-qing, HU Zi-yao, LEI Jian-feng, DAI Pei-hong, LIU Chao, DENG Jia-hui, LIU Min, SUN Ling, LIU Xiao-dong, LI Yue. Cloning and Functional Analysis of Gene GhTIFY9 Related to Cotton Verticillium Wilt Resistance [J]. Biotechnology Bulletin, 2022, 38(8): 127-134. |
| [12] | ZHAO Zeng-qiang, GUO Wen-ting, ZHANG Xi, LI Xiao-ling, ZHANG Wei. Cloning and Functional Analysis of GhERF5-4D Gene Related to Fusarium oxysporum Resistance in Cotton [J]. Biotechnology Bulletin, 2022, 38(4): 193-201. |
| [13] | ZHAO Yi, LEI Jian-feng, LIU Min, HU Zi-yao, DAI Pei-hong, LIU Chao, LI Yue, LIU Xiao-dong. Research on the Carrying Capacity of CLCrV-mediated VIGE System [J]. Biotechnology Bulletin, 2022, 38(11): 210-219. |
| [14] | HU Zi-yao, DAI Pei-hong, LIU Chao, Madina Mulati, WANG Qian, Wugalihan Abuduwili, ZHAO Yi, SUN Ling, XU Shi-jia, LI Yue. Molecular Cloning,Expression and VIGS Construction of a Small GTP-binding Protein Gene GhROP3 in Gossypium hirsutum [J]. Biotechnology Bulletin, 2021, 37(9): 106-113. |
| [15] | LIU Yuan-yuan, YANG Dong-jie, ZUO Dong-yun, CHENG Hai-liang, ZHANG You-ping, LV Li-min, WANG Qiao-lian, SONG Guo-li. Cloning and Functional Verification of GhD6PKL2 from Gossypium hirsutum [J]. Biotechnology Bulletin, 2021, 37(8): 111-120. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||