








Biotechnology Bulletin ›› 2023, Vol. 39 ›› Issue (7): 131-142.doi: 10.13560/j.cnki.biotech.bull.1985.2023-0015
Previous Articles Next Articles
WANG Shuai1( ), FENG Yu-mei2, BAI Miao1, DU Wei-jun1, YUE Ai-qin1(
), FENG Yu-mei2, BAI Miao1, DU Wei-jun1, YUE Ai-qin1( )
)
Received:2023-01-09
Online:2023-07-26
Published:2023-08-17
Contact:
YUE Ai-qin
E-mail:15234570975@163.com;yueaiqinnd@126.com
WANG Shuai, FENG Yu-mei, BAI Miao, DU Wei-jun, YUE Ai-qin. Functional Analysis of Soybean Gene GmHMGR Responding to Exogenous Hormones and Abiotic Stresses[J]. Biotechnology Bulletin, 2023, 39(7): 131-142.
| 基因Gene | 登录号Gene ID | 正向引物Forward primer(5'-3') | 反向引物Reverse primer(5'-3') |
|---|---|---|---|
| GmActin | XM_003552652 | GGTGGTTCTATCTTGGCATC | CTTTCGCTTCAATAACCCTA |
| qPCR-GmHMGR1 | XM_003517069.4 | GAAAAATCTTACTGGGTCTGCTATG | CTGATTGAGACGCCAGTTGTG |
| qPCR-GmHMGR2 | XM_014775260.2 | TGCTGGTTCTGCTGTCGC | CAAGTAAATTCAAGCAAGCAGATTA |
| qPCR-GmHMGR3 | XM_003537651.3 | TTTCTCCAGAGTGATTTTCC | CAGACCCAGTAAGGTTTTTC |
| qPCR-GmHMGR4 | XM_003534178.4 | TTCTTCAGAGTGACTTCC | GTCTTCAACACCTTCTTC |
| qPCR-GmHMGR5 | XM_006605513.3 | TTCTTCGGCATCCACTTCGTCCAG | GCTGTGCGTCTCCTTCTATGATGG |
| qPCR-GmHMGR6 | XM_003547838.4 | AAGCCGCAGCCGTGAATTGG | CCACCAAGAGCACCAGCCATG |
| qPCR-GmHMGR7 | XM_003519426.4 | TTCGGACAAGAAACCTGC | AAACCCACCAAGAGCACC |
| qPCR-GmHMGR8 | XM_003545508.4 | GCCGTTGTTGTTGGATGG | GCAATCCCTCAAAACCACA |
| AtActin | XM_021028231.1 | GGTAACATTGTGCTCAGTGGTGG | AACGACCTTAATCTTCATGCTGC |
| qPCR-AtDXS | NM_117647.3 | GAAGTCGCAAAGGGTATGACAAAGCA | CTGGATCAAATTTCACAACACCATGGTAT |
| qPCR-AtDXR | NM_125674.3 | TTCTGCCATATTTCAGTGTATTCAAGGTT | GCACAGATGAATCCTGTGTTTCAATC |
| qPCR-AtSQS1 | NM_119630.4 | GGAGAAGCAGATCCCTCCTG | AGTAGAACACACACGGCG |
| qPCR-AtSQS2 | NM_119631.3 | ATCTCAAAGCAAACCAATGTAAG | AGAACAACTGAAAAGGAAGAGAG |
| qPCR-AtCAS | NM_126681.3 | GACGGAGGTTGGGGTTTA | TGATTTAGTATCCAGTCTCGTCC |
| qPCR-AtIPI | NM_121649.6 | TGGGATCATGTTGAGAAAGGAAC | GTTGCCCAGTTTTGTCTGTAATCA |
Table 1 Primers sequences by real-time quantitative PCR
| 基因Gene | 登录号Gene ID | 正向引物Forward primer(5'-3') | 反向引物Reverse primer(5'-3') |
|---|---|---|---|
| GmActin | XM_003552652 | GGTGGTTCTATCTTGGCATC | CTTTCGCTTCAATAACCCTA |
| qPCR-GmHMGR1 | XM_003517069.4 | GAAAAATCTTACTGGGTCTGCTATG | CTGATTGAGACGCCAGTTGTG |
| qPCR-GmHMGR2 | XM_014775260.2 | TGCTGGTTCTGCTGTCGC | CAAGTAAATTCAAGCAAGCAGATTA |
| qPCR-GmHMGR3 | XM_003537651.3 | TTTCTCCAGAGTGATTTTCC | CAGACCCAGTAAGGTTTTTC |
| qPCR-GmHMGR4 | XM_003534178.4 | TTCTTCAGAGTGACTTCC | GTCTTCAACACCTTCTTC |
| qPCR-GmHMGR5 | XM_006605513.3 | TTCTTCGGCATCCACTTCGTCCAG | GCTGTGCGTCTCCTTCTATGATGG |
| qPCR-GmHMGR6 | XM_003547838.4 | AAGCCGCAGCCGTGAATTGG | CCACCAAGAGCACCAGCCATG |
| qPCR-GmHMGR7 | XM_003519426.4 | TTCGGACAAGAAACCTGC | AAACCCACCAAGAGCACC |
| qPCR-GmHMGR8 | XM_003545508.4 | GCCGTTGTTGTTGGATGG | GCAATCCCTCAAAACCACA |
| AtActin | XM_021028231.1 | GGTAACATTGTGCTCAGTGGTGG | AACGACCTTAATCTTCATGCTGC |
| qPCR-AtDXS | NM_117647.3 | GAAGTCGCAAAGGGTATGACAAAGCA | CTGGATCAAATTTCACAACACCATGGTAT |
| qPCR-AtDXR | NM_125674.3 | TTCTGCCATATTTCAGTGTATTCAAGGTT | GCACAGATGAATCCTGTGTTTCAATC |
| qPCR-AtSQS1 | NM_119630.4 | GGAGAAGCAGATCCCTCCTG | AGTAGAACACACACGGCG |
| qPCR-AtSQS2 | NM_119631.3 | ATCTCAAAGCAAACCAATGTAAG | AGAACAACTGAAAAGGAAGAGAG |
| qPCR-AtCAS | NM_126681.3 | GACGGAGGTTGGGGTTTA | TGATTTAGTATCCAGTCTCGTCC |
| qPCR-AtIPI | NM_121649.6 | TGGGATCATGTTGAGAAAGGAAC | GTTGCCCAGTTTTGTCTGTAATCA |

Fig. 2 Expression patterns of GmHMGR gene in response to hormone treatment in soybean A: MeJA treatment; B: ABA treatment.The data represent the mean ± SE of three biological replicates
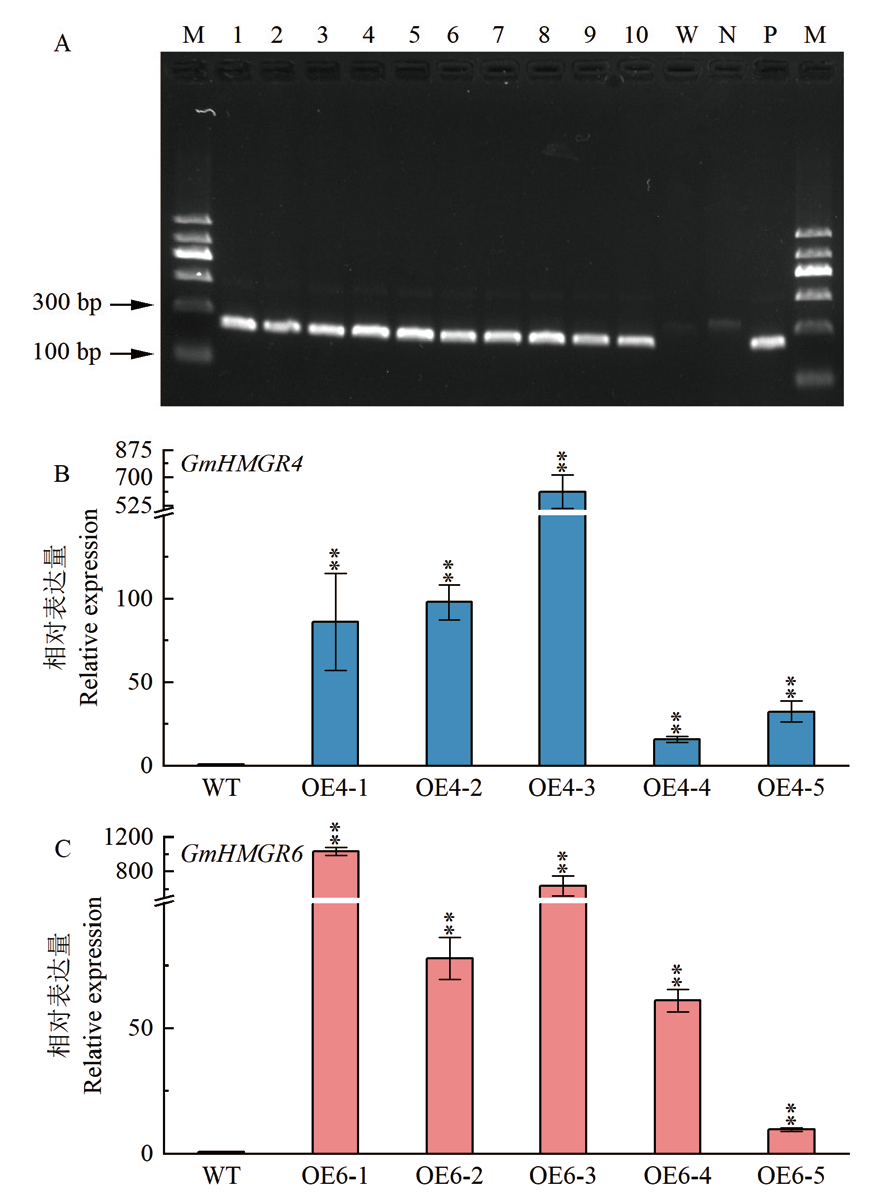
Fig. 4 Identification of T3 transgenic A. thaliana A: Identification of T3 generation A. thaliana by PCR; M: DNA molecular weight marker. Lane 1-5: GmHMGR4 transgenic lines; lane 6-10: GmHMGR6 transgenic lines; W: wild-type control; N: negative control; P: positive control; B: qPCR detection of overexpressed GmHMGR4 transgenic A. thaliana; C: qPCR detection of overexpressed GmHMGR6 transgenic A. thaliana. * indicates significant difference at 0.05 level, and ** indicates significant difference at 0.01 level. The same below

Fig. 6 Phenotypic analysis of transgenic A. thaliana after oxidative treatment and salt stress A: Growth phenotypes of wild-type and transgenic A. thaliana after oxidative and salt stress treatments. B: Size statistics of A. thaliana rosette diameter. C, D: Analysis of relative conductivity and leaf damage. E: Determination of MDA content
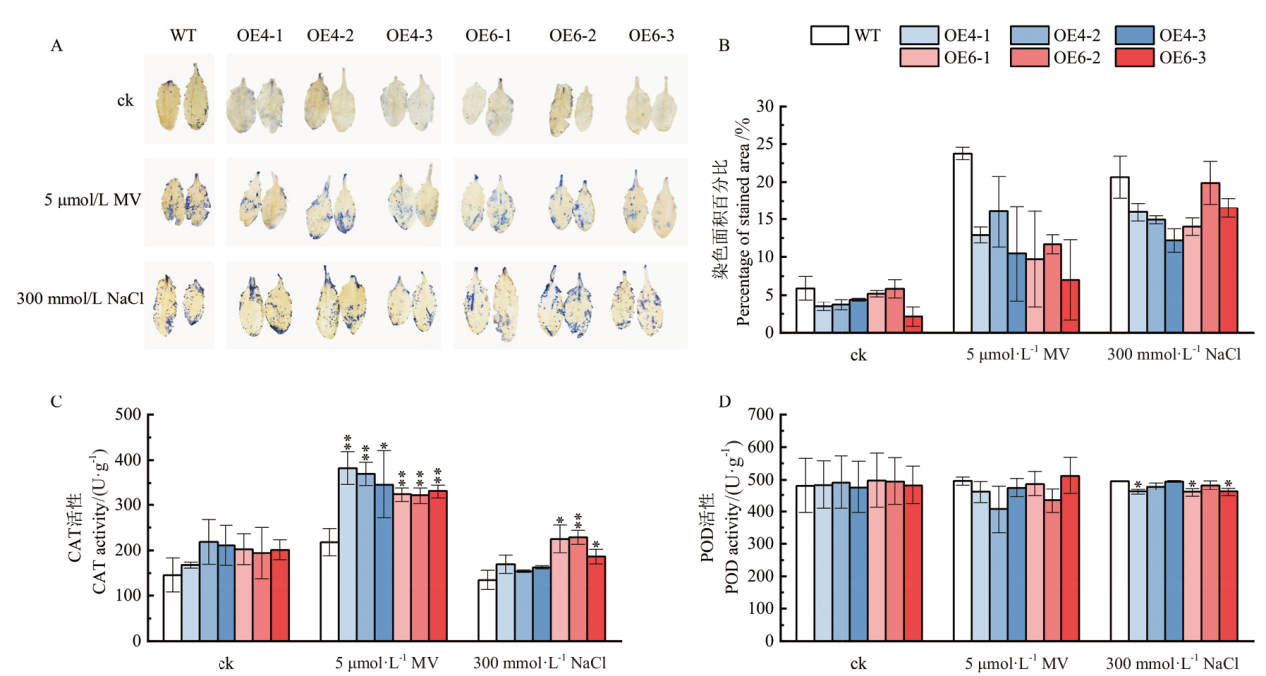
Fig. 7 Stress-resistant ability analysis of transgenic A. thaliana after oxidative treatment and salt stress A: NBT staining of A. thaliana leaves after oxidative and salt stress treatments. B: Statistics of stained area percentage. C, D: Determination of CAT and POD activities
| [1] | 马靓, 丁鹏, 杨广笑, 等. 植物类萜生物合成途径及关键酶的研究进展[J]. 生物技术通报, 2006(S1): 22-30. |
| Ma L, Ding P, Yang GX, et al. Advances on the plant terpenoid isoprenoid biosynthetic pathway and its key enzymes[J]. Biotechnol Bull, 2006(S1): 22-30. | |
| [2] |
Vranová E, Coman D, Gruissem W. Network analysis of the MVA and MEP pathways for isoprenoid synthesis[J]. Annu Rev Plant Biol, 2013, 64: 665-700.
doi: 10.1146/annurev-arplant-050312-120116 pmid: 23451776 |
| [3] |
Pulido P, Perello C, Rodriguez-Concepcion M. New insights into plant isoprenoid metabolism[J]. Mol Plant, 2012, 5(5): 964-967.
doi: 10.1093/mp/sss088 pmid: 22972017 |
| [4] |
Dong N, Ponciano G, McMahan CM, et al. Overexpression of 3-hydroxy-3-methylglutaryl coenzyme A reductase in Parthenium argentatum(guayule)[J]. Ind Crops Prod, 2013, 46: 15-24.
doi: 10.1016/j.indcrop.2012.12.044 URL |
| [5] |
Kirby J, Keasling JD. Biosynthesis of plant isoprenoids: perspectives for microbial engineering[J]. Annu Rev Plant Biol, 2009, 60: 335-355.
doi: 10.1146/annurev.arplant.043008.091955 pmid: 19575586 |
| [6] |
Bick JA, Lange BM. Metabolic cross talk between cytosolic and plastidial pathways of isoprenoid biosynthesis: unidirectional transport of intermediates across the chloroplast envelope membrane[J]. Arch Biochem Biophys, 2003, 415(2): 146-154.
doi: 10.1016/s0003-9861(03)00233-9 pmid: 12831836 |
| [7] |
陈妤, 朱沛煌, 李荣, 等. 植物异戊烯基转移酶研究进展[J]. 生物技术通报, 2021, 37(2): 149-161.
doi: 10.13560/j.cnki.biotech.bull.1985.2020-0476 URL |
|
Chen Y, Zhu PH, Li R, et al. Research progress of plant prenyltransferases[J]. Biotechnol Bull, 2021, 37(2): 149-161.
doi: 10.13560/j.cnki.biotech.bull.1985.2020-0476 URL |
|
| [8] |
Suzuki M, Kamide Y, Nagata N, et al. Loss of function of 3-hydroxy-3-methylglutaryl coenzyme A reductase 1(HMG1)in Arabidopsis leads to dwarfing, early senescence and male sterility, and reduced sterol levels[J]. Plant J, 2004, 37(5): 750-761.
doi: 10.1111/tpj.2004.37.issue-5 URL |
| [9] |
Rao S, Meng XX, Liao YL, et al. Characterization and functional analysis of two novel 3-hydroxy-3-methylglutaryl-coenzyme A reductase genes(GbHMGR2 and GbHMGR3)from Ginkgo biloba[J]. Sci Rep, 2019, 9(1): 14109.
doi: 10.1038/s41598-019-50629-8 |
| [10] | 郝金凤, 李彤彤, 郭艳琼, 等. 甜瓜HMGR基因全长cDNA的克隆及序列分析[J]. 生物技术通报, 2010(8): 106-109, 115. |
| Hao JF, Li TT, Guo YQ, et al. Cloning and sequence analysis of the 3-hydroxy-3-methylglutaryl coenzyme-a reductase gene cDNA from melon cultivar Hetao(Cucumis melo L.cv.Hetao)[J]. Biotechnol Bull, 2010(8): 106-109, 115. | |
| [11] |
Liu W, Zhang ZQ, Li W, et al. Genome-wide identification and comparative analysis of the 3-hydroxy-3-methylglutaryl coenzyme a reductase(HMGR)gene family in Gossypium[J]. Molecules, 2018, 23(2): 193.
doi: 10.3390/molecules23020193 URL |
| [12] |
Kim YJ, Lee OR, Oh JY, et al. Functional analysis of 3-hydroxy-3-methylglutaryl coenzyme a reductase encoding genes in triterpene saponin-producing ginseng[J]. Plant Physiol, 2014, 165(1): 373-387.
doi: 10.1104/pp.113.222596 URL |
| [13] |
Zhang M, Liu H, Wang Q, et al. The 3-hydroxy-3-methylglutaryl-coenzyme A reductase 5 gene from Malus domestica enhances oxidative stress tolerance in Arabidopsis thaliana[J]. Plant Physiol Biochem, 2020, 146: 269-277.
doi: 10.1016/j.plaphy.2019.11.031 URL |
| [14] |
Choi D, Ward BL, Bostock RM. Differential induction and suppression of potato 3-hydroxy-3-methylglutaryl coenzyme A reductase genes in response to Phytophthora infestans and to its elicitor arachidonic acid[J]. Plant Cell, 1992, 4(10): 1333-1344.
pmid: 1283354 |
| [15] |
Lumbreras V, Campos N, Boronat A. The use of an alternative promoter in the Arabidopsis thaliana HMG1 gene generates an mRNA that encodes a novel 3-hydroxy-3-methylglutaryl coenzyme A reductase isoform with an extended N-terminal region[J]. Plant J, 1995, 8(4): 541-549.
doi: 10.1046/j.1365-313x.1995.8040541.x pmid: 7496400 |
| [16] |
Rupasinghe HPV, Almquist KC, Paliyath G, et al. Cloning of hmg1 and hmg2 cDNAs encoding 3-hydroxy-3-methylglutaryl coenzyme A reductase and their expression and activity in relation to α-farnesene synthesis in apple[J]. Plant Physiol Biochem, 2001, 39(11): 933-947.
doi: 10.1016/S0981-9428(01)01316-X URL |
| [17] | 陈俞裴, 申甜, 邱飞, 等. 青蒿HMGR基因家族克隆及对茉莉酸甲酯和损伤的响应[J]. 西南大学学报: 自然科学版, 2017, 39(12): 44-51. |
| Chen YP, Shen T, Qiu F, et al. Molecular cloning and expression analysis of HMG-CoA(3-hydroxy-3-methylglutaryl-CoA)reductase gene family upon MeJA and mechanical injury treatments in Artemisia annua L[J]. J Southwest Univ Nat Sci, 2017, 39(12): 44-51. | |
| [18] | 刁秀楠, 斛如媛, 高春艳, 等. 大豆HMGR基因家族的鉴定与表达分析[J]. 山西农业大学学报: 自然科学版, 2019, 39(4)9-16 |
| Diao XN, Hu RY, Gao CY, et al. Identification and expression analysis of HMGR gene family in Glycine max[J]. J Shanxi Agric Univ Nat Sci Ed, 2019, 39(4)9-16 | |
| [19] | Abramoff M, Magalhães P, Ram SJ. Image processing with ImageJ[J]. Biophotonics Int, 2003, 11: 36-42. |
| [20] |
Bouvier F, Rahier A, Camara B. Biogenesis, molecular regulation and function of plant isoprenoids[J]. Prog Lipid Res, 2005, 44(6): 357-429.
doi: 10.1016/j.plipres.2005.09.003 pmid: 16289312 |
| [21] |
Jin HN, Song ZH, Nikolau BJ. Reverse genetic characterization of two paralogous acetoacetyl CoA thiolase genes in Arabidopsis reveals their importance in plant growth and development[J]. Plant J, 2012, 70(6): 1015-1032.
doi: 10.1111/tpj.2012.70.issue-6 URL |
| [22] |
Liao P, Chen XJ, Wang MF, et al. Improved fruit α-tocopherol, carotenoid, squalene and phytosterol contents through manipulation of Brassica juncea 3-HYDROXY-3-METHYLGLUTARYL-COA SYNTHASE1 in transgenic tomato[J]. Plant Biotechnol J, 2018, 16(3): 784-796.
doi: 10.1111/pbi.12828 pmid: 28881416 |
| [23] | Liao P, Wang H, Wang MF, et al. Transgenic tobacco overexpressing Brassica juncea HMG-CoA synthase 1 shows increased plant growth, pod size and seed yield[J]. PLoS One, 2014, 9(5): e98264. |
| [24] |
Manzano D, Andrade P, Caudepón D, et al. Suppressing farnesyl diphosphate synthase alters chloroplast development and triggers sterol-dependent induction of jasmonate- and Fe-related responses[J]. Plant Physiol, 2016, 172(1): 93-117.
doi: 10.1104/pp.16.00431 pmid: 27382138 |
| [25] |
Kevei Z, Lougnon G, Mergaert P, et al. 3-hydroxy-3-methylglutaryl coenzyme a reductase 1 interacts with NORK and is crucial for nodulation in Medicago truncatula[J]. Plant Cell, 2007, 19(12): 3974-3989.
doi: 10.1105/tpc.107.053975 URL |
| [26] | 黄晶, 段续伟, 张文娜, 等. 杜梨HMGR基因克隆及其转基因烟草种子耐盐性分析[J]. 中国农业大学学报, 2015, 20(1): 60-67. |
| Huang J, Duan XW, Zhang WN, et al. Clone of pear HMGR gene and salt-tolerance analysis of its transgenic tobacco seed[J]. J China Agric Univ, 2015, 20(1): 60-67. | |
| [27] |
Lv DM, Zhang TT, Deng S, et al. Functional analysis of the Malus domestica MdHMGR2 gene promoter in transgenic Arabidopsis thaliana[J]. Biol Plant, 2016, 60(4): 667-676.
doi: 10.1007/s10535-016-0637-z URL |
| [28] |
Lv DM, Zhang YH. Isolation and functional analysis of apple MdHMGR1 and MdHMGR4 gene promoters in transgenic Arabi-dopsis thaliana[J]. Plant Cell Tiss Organ Cult, 2017, 129(1): 133-143.
doi: 10.1007/s11240-016-1162-7 URL |
| [29] |
Leivar P, Antolín-Llovera M, Ferrero S, et al. Multilevel control of Arabidopsis 3-hydroxy-3-methylglutaryl coenzyme A reductase by protein phosphatase 2A[J]. Plant Cell, 2011, 23(4): 1494-1511.
doi: 10.1105/tpc.110.074278 URL |
| [30] |
Lange I, Poirier BC, Herron BK, et al. Comprehensive assessment of transcriptional regulation facilitates metabolic engineering of isoprenoid accumulation in Arabidopsis[J]. Plant Physiol, 2015, 169(3): 1595-1606.
doi: 10.1104/pp.15.00573 pmid: 26282236 |
| [31] |
Nieto B, Forés O, Arró M, et al. Arabidopsis 3-hydroxy-3-methylglutaryl-CoA reductase is regulated at the post-translational level in response to alterations of the sphingolipid and the sterol biosynthetic pathways[J]. Phytochemistry, 2009, 70(1): 53-59.
doi: 10.1016/j.phytochem.2008.10.010 URL |
| [32] | Dai ZB, Cui GH, Zhou SF, et al. Cloning and characterization of a novel 3-hydroxy-3-methylglutaryl coenzyme A reductase gene from Salvia miltiorrhiza involved in diterpenoid tanshinone accumulation[J]. J Plant Physiol, 2011, 168(2): 148-157. |
| [33] |
Holmberg N, Harker M, Wallace AD, et al. Co-expression of N-terminal truncated 3-hydroxy-3-methylglutaryl CoA reductase and C24-sterol methyltransferase type 1 in transgenic tobacco enhances carbon flux towards end-product sterols[J]. Plant J, 2003, 36(1): 12-20.
pmid: 12974807 |
| [34] |
Laule O, Fürholz A, Chang HS, et al. Crosstalk between cytosolic and plastidial pathways of isoprenoid biosynthesis in Arabidopsis thaliana[J]. Proc Natl Acad Sci USA, 2003, 100(11): 6866-6871.
doi: 10.1073/pnas.1031755100 URL |
| [35] | Pallavi S, Bhushan JA, Shanker DR, et al. Reactive oxygen species, oxidative damage, and antioxidative defense mechanism in plants under stressful conditions[J]. J Bot, 2012, 2012: 1-26. |
| [36] |
Wei H, Movahedi A, Xu C, et al. Overexpression of PtHMGR enhances drought and salt tolerance of poplar[J]. Ann Bot, 2020, 125(5): 785-803.
doi: 10.1093/aob/mcz158 URL |
| [37] |
Wang H, Nagegowda DA, Rawat R, et al. Overexpression of Brassica juncea wild-type and mutant HMG-CoA synthase 1 in Arabidopsis up-regulates genes in sterol biosynthesis and enhances sterol production and stress tolerance[J]. Plant Biotechnol J, 2012, 10(1): 31-42.
doi: 10.1111/j.1467-7652.2011.00631.x pmid: 21645203 |
| [38] |
Vickers CE, Gershenzon J, Lerdau MT, et al. A unified mechanism of action for volatile isoprenoids in plant abiotic stress[J]. Nat Chem Biol, 2009, 5(5): 283-291.
doi: 10.1038/nchembio.158 pmid: 19377454 |
| [39] |
Loreto F, Schnitzler JP. Abiotic stresses and induced BVOCs[J]. Trends Plant Sci, 2010, 15(3): 154-166.
doi: 10.1016/j.tplants.2009.12.006 pmid: 20133178 |
| [40] |
Loreto F, Dicke M, Schnitzler JP, et al. Plant volatiles and the environment[J]. Plant Cell Environ, 2014, 37(8): 1905-1908.
doi: 10.1111/pce.12369 URL |
| [1] | KANG Ling-yun, HAN Lu-lu, HAN De-ping, CHEN Jian-sheng, GAN Han-ling, XING Kai, MA You-ji, CUI Kai. Effect of Melatonin on Protecting the Jejunum Mucosal Epithelial Cells from Oxidative Stress Damage [J]. Biotechnology Bulletin, 2023, 39(9): 291-299. |
| [2] | LI Wen-chen, LIU Xin, KANG Yue, LI Wei, QI Ze-zheng, YU Lu, WANG Fang. Optimization and Application of Tobacco Rattle Virus-induced Gene Silencing System in Soybean [J]. Biotechnology Bulletin, 2023, 39(7): 143-150. |
| [3] | WEI Xi-ya, QIN Zhong-wei, LIANG La-mei, LIN Xin-qi, LI Ying-zhi. Mechanism of Melatonin Seed Priming in Improving Salt Tolerance of Capsicum annuum [J]. Biotechnology Bulletin, 2023, 39(7): 160-172. |
| [4] | ZHAI Ying, LI Ming-yang, ZHANG Jun, ZHAO Xu, YU Hai-wei, LI Shan-shan, ZHAO Yan, ZHANG Mei-juan, SUN Tian-guo. Heterologous Expression of Soybean Transcription Factor GmNF-YA19 Improves Drought Resistance of Transgenic Tobacco [J]. Biotechnology Bulletin, 2023, 39(5): 224-232. |
| [5] | MA Yu-qian, SUN Dong-hui, YUE Hao-feng, XIN Jia-yu, LIU Ning, CAO Zhi-yan. Identification, Heterologous Expression and Functional Analysis of a GH61 Family Glycoside Hydrolase from Setosphaeria turcica with the Assisting Function in Degrading Cellulose [J]. Biotechnology Bulletin, 2023, 39(4): 124-135. |
| [6] | CHEN Nan-nan, WANG Chun-lai, JIANG Zhen-zhong, JIAO Peng, GUAN Shu-yan, MA Yi-yong. Genetic Transformation and Chilling Resistance Analysis of Maize ZmDHN15 Gene in Tobacco [J]. Biotechnology Bulletin, 2023, 39(4): 259-267. |
| [7] | HOU Xiao-yuan, CHE Zheng-zheng, LI Heng-jing, DU Chong-yu, XU Qian, WANG Qun-qing. Construction of the Soybean Membrane System cDNA Library and Interaction Proteins Screening for Effector PsAvr3a [J]. Biotechnology Bulletin, 2023, 39(4): 268-276. |
| [8] | WANG Hai-long, LI Yu-qian, WANG Bo, XING Guo-fang, ZHANG Jie-wei. Isolation and Expression Analysis of SiMAPK3 in Setaria italica L. [J]. Biotechnology Bulletin, 2023, 39(3): 123-132. |
| [9] | YANG Chun-hong, DONG Lu, CHEN Lin, SONG Li. Characterization of Soybean VAS1 Gene Family and Its Involvement in Lateral Root Development [J]. Biotechnology Bulletin, 2023, 39(3): 133-142. |
| [10] | WANG Qi, HU Zhe, FU Wei, LI Guang-zhe, HAO Lin. Regulation of Burkholderia sp. GD17 on the Drought Tolerance of Cucumber Seedlings [J]. Biotechnology Bulletin, 2023, 39(3): 163-175. |
| [11] | DU Qing-jie, ZHOU Lu-yao, YANG Si-zhen, ZHANG Jia-xin, CHEN Chun-lin, LI Juan-qi, LI Meng, ZHAO Shi-wen, XIAO Huai-juan, WANG Ji-qing. Overexpression of CaCP1 Enhances Salt Stress Sensibility in Transgenic Tobacco [J]. Biotechnology Bulletin, 2023, 39(2): 172-182. |
| [12] | CHEN Yi-bo, YANG Wan-ming, YUE Ai-qin, WANG Li-xiang, DU Wei-jun, WANG Min. Construction of Soybean Genetic Map Based on SLAF Markers and QTL Mapping Analysis of Salt Tolerance at Seedling Stage [J]. Biotechnology Bulletin, 2023, 39(2): 70-79. |
| [13] | MIAO Shu-nan, GAO Yu, LI Xin-ru, CAI Gui-ping, ZHANG Fei, XUE Jin-ai, JI Chun-li, LI Run-zhi. Functional Analysis of Soybean GmPDAT1 Genes in the Oil Biosynthesis and Response to Abiotic Stresses [J]. Biotechnology Bulletin, 2023, 39(2): 96-106. |
| [14] | YAN Xiong-ying, WANG Zhen, WANG Xia, YANG Shi-hui. Microbial Sulfur Metabolism and Stress Resistance [J]. Biotechnology Bulletin, 2023, 39(11): 150-167. |
| [15] | WANG Ming-tao, LIU Jian-wei, ZHAO Chun-zhao. Molecular Mechanisms of Cell Wall Integrity in Plants Under Salt Stress [J]. Biotechnology Bulletin, 2023, 39(11): 18-27. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||