








Biotechnology Bulletin ›› 2023, Vol. 39 ›› Issue (8): 148-158.doi: 10.13560/j.cnki.biotech.bull.1985.2022-1534
Previous Articles Next Articles
CHEN Xiao-ling1( ), LIAO Dong-qing2, HUANG Shang-fei1, CHEN Ying1, LU Zhi-long1, CHEN Dong1
), LIAO Dong-qing2, HUANG Shang-fei1, CHEN Ying1, LU Zhi-long1, CHEN Dong1
Received:2022-12-18
Online:2023-08-26
Published:2023-09-05
Contact:
CHEN Xiao-ling
E-mail:xiaolingchen1116@163.com
CHEN Xiao-ling, LIAO Dong-qing, HUANG Shang-fei, CHEN Ying, LU Zhi-long, CHEN Dong. Advances in CRISPR/Cas9 System Modifying Saccharomycescerevisiae[J]. Biotechnology Bulletin, 2023, 39(8): 148-158.
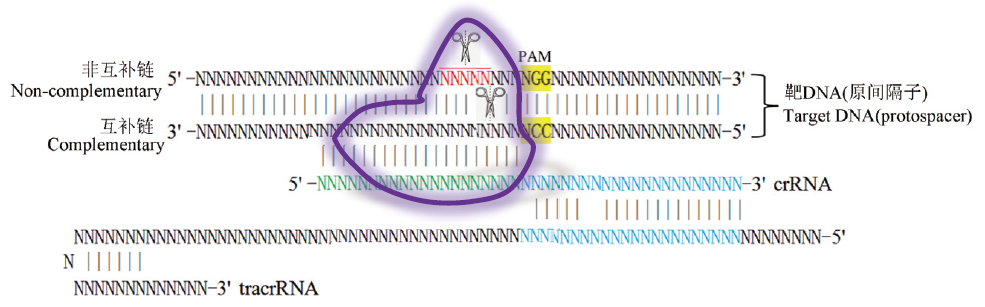
Site-specific DNA cleavage by CRISPR/Cas9 system Case N indicates base A, C, G,T(DNA)or U(RNA); indicate Cas9 whose cleavage sites are marked with scissors(N in red show the probely cut sits). Regions of crRNA complementarity to tracrRNA(blue), protospacer DNA(green)and PAM sequence(in yellow background)are represented
| 名称 Name | 链接 Link | 物种Species | 优点 Advantage | 缺点 Disadvantage | 参考文献Reference |
|---|---|---|---|---|---|
| GuideScan | - | 任何 微生物 | 能以目标酿酒酵母的基因组为参考基因组、有助于减少脱靶位点,设计的靶序列比the mit.edu web interface[ | 需在本地计算机安装,需使用命令行,需提供参考基因组 | [ |
| Yeastriction | http://yeastriction.tnw.tudelft.nl/ | 酿酒酵母 | 设计的靶序列比ChopChop[ | 容易脱靶(除非参考基因组是目标酿酒酵母的基因组) | [ |
| CRISPy | http://staff.biosustain.dtu.dk/laeb/crispy_yeast/ | 酿酒酵母 | 不需上传参考基因组 | 只能以酿酒酵母S288C的基因组为参考基因组,为其他酿酒酵母设计靶序列时,容易脱靶 | [ |
| CRISPy-web | http://crispy.secondarymetabolites.org/ | 任何 微生物 | 免安装,不需使用命令行,能以目标酿酒酵母的基因组为参考基因组、有助于减少脱靶位点 | 需上传参考基因组 | [ |
Table 1 SgRNA design tools available for S. cerevisiae
| 名称 Name | 链接 Link | 物种Species | 优点 Advantage | 缺点 Disadvantage | 参考文献Reference |
|---|---|---|---|---|---|
| GuideScan | - | 任何 微生物 | 能以目标酿酒酵母的基因组为参考基因组、有助于减少脱靶位点,设计的靶序列比the mit.edu web interface[ | 需在本地计算机安装,需使用命令行,需提供参考基因组 | [ |
| Yeastriction | http://yeastriction.tnw.tudelft.nl/ | 酿酒酵母 | 设计的靶序列比ChopChop[ | 容易脱靶(除非参考基因组是目标酿酒酵母的基因组) | [ |
| CRISPy | http://staff.biosustain.dtu.dk/laeb/crispy_yeast/ | 酿酒酵母 | 不需上传参考基因组 | 只能以酿酒酵母S288C的基因组为参考基因组,为其他酿酒酵母设计靶序列时,容易脱靶 | [ |
| CRISPy-web | http://crispy.secondarymetabolites.org/ | 任何 微生物 | 免安装,不需使用命令行,能以目标酿酒酵母的基因组为参考基因组、有助于减少脱靶位点 | 需上传参考基因组 | [ |
| [1] |
Caspeta L, Chen Y, Ghiaci P, et al. Biofuels. Altered sterol composition renders yeast thermotolerant[J]. Science, 2014, 346(6205):75-78.
doi: 10.1126/science.1258137 pmid: 25278608 |
| [2] |
Lu ZL, Wu YL, Chen Y, et al. Role of spt23 in Saccharomyces cerevisiae thermal tolerance[J]. Appl Microbiol Biotechnol, 2022, 106(9): 3691-3705.
doi: 10.1007/s00253-022-11920-3 |
| [3] |
Chen Y, Lu ZL, Chen D, et al. Transcriptomic analysis and driver mutant prioritization for differentially expressed genes from a Saccharomyces cerevisiae strain with high glucose tolerance generated by UV irradiation[J]. RSC Adv, 2017, 7(62): 38784-38797.
doi: 10.1039/C7RA06146C URL |
| [4] | 吴仁智, 陈东, 关妮, 等. 一种高产乙醇发酵菌株及其诱变育种方法:CN114317297B[P]. 2022-11-08. |
| Wu RZ, Chen D, Guan N, et al. A high-yielding ethanol-producing yeast strain and its mutation breeding method: CN114317297B[P]. 2022-11-08. | |
| [5] |
Chen XL, Lu ZL, Chen Y, et al. Deletion of the MBP1 gene, involved in the cell cycle, affects respiration and pseudohyphal differentiation in Saccharomyces cerevisiae[J]. Microbiol Spectr, 2021, 9(1): e0008821.
doi: 10.1128/Spectrum.00088-21 URL |
| [6] |
Wu RZ, Chen D, Cao SW, et al. Enhanced ethanol production from sugarcane molasses by industrially engineered Saccharomyces cerevisiae via replacement of the PHO4 gene[J]. RSC Adv, 2020, 10(4): 2267-2276.
doi: 10.1039/C9RA08673K URL |
| [7] | 陈英, 陈东, 陆琦, 等. 一株高产甘蔗糖蜜酒精的基因重组酿酒酵母:CN105670953A[P]. 2016-06-15. |
| Chen Y, Chen D, Lu Q, et al. A recombinant Saccharomyces cerevisiae with high production of ethanol from sugarcane molasses: CN105670953A[P]. 2016-06-15. | |
| [8] | 吴仁智, 陈东, 曹树威, 等. 一种用于促进高产甘蔗糖蜜酒精发酵的菌株及其应用:CN110904123B[P]. 2021-11-05. |
| Wu R, Chen D, Cao S, et al. A strain for enhancing ethanol production from sugarcane molasses and its application: CN110904123B[P]. 2021-11-05. | |
| [9] | 方佩佩, 王世清, 李静, 等. 耐酒精酿酒酵母大气压等离子体诱变条件的建立及选育[J]. 酿酒科技, 2016(9): 31-37. |
| Fang PP, Wang SQ, Li J, et al. Establishment of the conditions of atmospheric plasma-inducing mutation of S. cerevisiae and breeding of an ethanol-tolerant strain[J]. Liquor Mak Sci Technol, 2016(9): 31-37. | |
| [10] |
Wiedenheft B, Sternberg SH, Doudna JA. RNA-guided genetic silencing systems in bacteria and Archaea[J]. Nature, 2012, 482(7385): 331-338.
doi: 10.1038/nature10886 |
| [11] |
Bhaya D, Davison M, Barrangou R. CRISPR-Cas systems in bacteria and Archaea: versatile small RNAs for adaptive defense and regulation[J]. Annu Rev Genet, 2011, 45: 273-297.
doi: 10.1146/annurev-genet-110410-132430 pmid: 22060043 |
| [12] |
Terns MP, Terns RM. CRISPR-based adaptive immune systems[J]. Curr Opin Microbiol, 2011, 14(3): 321-327.
doi: 10.1016/j.mib.2011.03.005 pmid: 21531607 |
| [13] |
Deltcheva E, Chylinski K, Sharma CM, et al. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III[J]. Nature, 2011, 471(7340): 602-607.
doi: 10.1038/nature09886 |
| [14] |
Haurwitz RE, Jinek M, Wiedenheft B, et al. Sequence- and structure-specific RNA processing by a CRISPR endonuclease[J]. Science, 2010, 329(5997): 1355-1358.
doi: 10.1126/science.1192272 pmid: 20829488 |
| [15] |
Brouns SJJ, Jore MM, Lundgren M, et al. Small CRISPR RNAs guide antiviral defense in prokaryotes[J]. Science, 2008, 321(5891): 960-964.
doi: 10.1126/science.1159689 pmid: 18703739 |
| [16] |
Wiedenheft B, Lander GC, Zhou KH, et al. Structures of the RNA-guided surveillance complex from a bacterial immune system[J]. Nature, 2011, 477(7365): 486-489.
doi: 10.1038/nature10402 |
| [17] |
Makarova KS, Haft DH, Barrangou R, et al. Evolution and classification of the CRISPR-Cas systems[J]. Nat Rev Microbiol, 2011, 9(6): 467-477.
doi: 10.1038/nrmicro2577 pmid: 21552286 |
| [18] |
Makarova KS, Aravind L, Wolf YI, et al. Unification of Cas protein families and a simple scenario for the origin and evolution of CRISPR-Cas systems[J]. Biol Direct, 2011, 6: 38.
doi: 10.1186/1745-6150-6-38 pmid: 21756346 |
| [19] |
Horvath P, Barrangou R. CRISPR/cas, the immune system of bacteria and Archaea[J]. Science, 2010, 327(5962): 167-170.
doi: 10.1126/science.1179555 pmid: 20056882 |
| [20] |
Jinek M, Chylinski K, Fonfara I, et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity[J]. Science, 2012, 337(6096): 816-821.
doi: 10.1126/science.1225829 pmid: 22745249 |
| [21] |
Jacobus AP, Barreto JA, de Bem LS, et al. EasyGuide plasmids support in vivo assembly of gRNAs for CRISPR/Cas9 applications in Saccharomyces cerevisiae[J]. ACS Synth Biol, 2022, 11(11): 3886-3891.
doi: 10.1021/acssynbio.2c00348 pmid: 36257021 |
| [22] |
Yang PZ, Jiang SY, Jiang SW, et al. CRISPR-Cas9 approach constructed engineered Saccharomyces cerevisiae with the deletion of GPD2, FPS1, and ADH2 to enhance the production of ethanol[J]. JoF, 2022, 8(7): 703.
doi: 10.3390/jof8070703 URL |
| [23] |
Yang PZ, Jiang SY, Lu SH, et al. Ethanol yield improvement in Saccharomyces cerevisiae GPD2 Delta FPS1 Delta ADH2 Delta DLD3 Delta mutant and molecular mechanism exploration based on the metabolic flux and transcriptomics approaches[J]. Microb Cell Fact, 2022, 21(1): 160.
doi: 10.1186/s12934-022-01885-3 |
| [24] |
Meng J, Qiu Y, Shi SB. CRISPR/Cas9 systems for the development of Saccharomyces cerevisiae cell factories[J]. Front Bioeng Biotechnol, 2020, 8: 594347.
doi: 10.3389/fbioe.2020.594347 URL |
| [25] |
Patra P, Das M, Kundu P, et al. Recent advances in systems and synthetic biology approaches for developing novel cell-factories in non-conventional yeasts[J]. Biotechnol Adv, 2021, 47: 107695.
doi: 10.1016/j.biotechadv.2021.107695 URL |
| [26] | Mans R, van Rossum HM, Wijsman M, et al. CRISPR/Cas9: a molecular Swiss army knife for simultaneous introduction of multiple genetic modifications in Saccharomyces cerevisiae[J]. FEMS Yeast Res, 2015, 15(2): fov004. |
| [27] |
Zhang GC, Kong II, Kim H, et al. Construction of a quadruple auxotrophic mutant of an industrial polyploid Saccharomyces cerevisiae strain by using RNA-guided Cas9 nuclease[J]. Appl Environ Microbiol, 2014, 80(24): 7694-7701.
doi: 10.1128/AEM.02310-14 URL |
| [28] |
Jakočiūnas T, Bonde I, Herrgård M, et al. Multiplex metabolic pathway engineering using CRISPR/Cas9 in Saccharomyces cerevisiae[J]. Metab Eng, 2015, 28: 213-222.
doi: S1096-7176(15)00010-5 pmid: 25638686 |
| [29] |
DiCarlo JE, Norville JE, Mali P, et al. Genome engineering in Saccharomyces cerevisiae using CRISPR-Cas systems[J]. Nucleic Acids Res, 2013, 41(7): 4336-4343.
doi: 10.1093/nar/gkt135 pmid: 23460208 |
| [30] |
Hsu PD, Scott DA, Weinstein JA, et al. DNA targeting specificity of RNA-guided Cas9 nucleases[J]. Nat Biotechnol, 2013, 31(9): 827-832.
doi: 10.1038/nbt.2647 pmid: 23873081 |
| [31] | Rainha J, Rodrigues JL, Rodrigues LR. CRISPR-Cas9: a powerful tool to efficiently engineer Saccharomyces cerevisiae[J]. Life(Basel), 2020, 11(1): 13. |
| [32] |
Xie KB, Minkenberg B, Yang YN. Boosting CRISPR/Cas9 multiplex editing capability with the endogenous tRNA-processing system[J]. Proc Natl Acad Sci USA, 2015, 112(11): 3570-3575.
doi: 10.1073/pnas.1420294112 pmid: 25733849 |
| [33] |
Port F, Bullock SL. Augmenting CRISPR applications in Drosophila with tRNA-flanked sgRNAs[J]. Nat Methods, 2016, 13(10): 852-854.
doi: 10.1038/nmeth.3972 |
| [34] |
Schwartz CM, Hussain MS, Blenner M, et al. Synthetic RNA polymerase III promoters facilitate high-efficiency CRISPR-Cas9-mediated genome editing in Yarrowia lipolytica[J]. ACS Synth Biol, 2016, 5(4): 356-359.
doi: 10.1021/acssynbio.5b00162 pmid: 26714206 |
| [35] | Xu L, Zhao LX, Gao YD, et al. Empower multiplex cell and tissue-specific CRISPR-mediated gene manipulation with self-cleaving ribozymes and tRNA[J]. Nucleic Acids Res, 2017, 45(5): e28. |
| [36] |
Ding D, Chen KY, Chen YD, et al. Engineering introns to express RNA guides for Cas9- and Cpf1-mediated multiplex genome editing[J]. Mol Plant, 2018, 11(4): 542-552.
doi: S1674-2052(18)30055-8 pmid: 29462720 |
| [37] |
Qi WW, Zhu T, Tian ZR, et al. High-efficiency CRISPR/Cas9 multiplex gene editing using the glycine tRNA-processing system-based strategy in maize[J]. BMC Biotechnol, 2016, 16(1): 58.
doi: 10.1186/s12896-016-0289-2 pmid: 27515683 |
| [38] |
Mefferd AL, Kornepati AVR, Bogerd HP, et al. Expression of CRISPR/Cas single guide RNAs using small tRNA promoters[J]. RNA, 2015, 21(9): 1683-1689.
doi: 10.1261/rna.051631.115 pmid: 26187160 |
| [39] |
Deaner M, Mejia J, Alper HS. Enabling graded and large-scale multiplex of desired genes using a dual-mode dCas9 activator in Saccharomyces cerevisiae[J]. ACS Synth Biol, 2017, 6(10): 1931-1943.
doi: 10.1021/acssynbio.7b00163 URL |
| [40] | Deaner M, Holzman A, Alper HS. Modular ligation extension of guide RNA operons(LEGO)for multiplexed dCas9 regulation of metabolic pathways in Saccharomyces cerevisiae[J]. Biotechnol J, 2018, 13(9): e1700582. |
| [41] |
Generoso WC, Gottardi M, Oreb M, et al. Simplified CRISPR-cas genome editing for Saccharomyces cerevisiae[J]. J Microbiol Methods, 2016, 127: 203-205.
doi: S0167-7012(16)30149-X pmid: 27327211 |
| [42] |
Moreno-Mateos MA, Vejnar CE, Beaudoin JD, et al. CRISPRscan: designing highly efficient sgRNAs for CRISPR-Cas9 targeting in vivo[J]. Nat Methods, 2015, 12(10): 982-988.
doi: 10.1038/nmeth.3543 pmid: 26322839 |
| [43] |
Fu YF, Sander JD, Reyon D, et al. Improving CRISPR-Cas nuclease specificity using truncated guide RNAs[J]. Nat Biotechnol, 2014, 32(3): 279-284.
doi: 10.1038/nbt.2808 pmid: 24463574 |
| [44] |
Sashital DG, Wiedenheft B, Doudna JA. Mechanism of foreign DNA selection in a bacterial adaptive immune system[J]. Mol Cell, 2012, 46(5): 606-615.
doi: 10.1016/j.molcel.2012.03.020 pmid: 22521690 |
| [45] |
Perez AR, Pritykin Y, Vidigal JA, et al. GuideScan software for improved single and paired CRISPR guide RNA design[J]. Nat Biotechnol, 2017, 35(4): 347-349.
doi: 10.1038/nbt.3804 pmid: 28263296 |
| [46] | Mojica FJM, Díez-Villaseñor C, García-Martínez J, et al. Short motif sequences determine the targets of the prokaryotic CRISPR defence system[J]. Microbiology(Reading), 2009, 155(Pt 3): 733-740. |
| [47] |
Christie KA, Guo JA, Silverstein RA, et al. Precise DNA cleavage using CRISPR-SpRYgests[J]. Nat Biotechnol, 2023, 41(3): 409-416.
doi: 10.1038/s41587-022-01492-y |
| [48] |
Walton RT, Christie KA, Whittaker MN, et al. Unconstrained genome targeting with near-PAMless engineered CRISPR-Cas9 variants[J]. Science, 2020, 368(6488): 290-296.
doi: 10.1126/science.aba8853 pmid: 32217751 |
| [49] |
Jiang WY, Bikard D, Cox D, et al. RNA-guided editing of bacterial genomes using CRISPR-Cas systems[J]. Nat Biotechnol, 2013, 31(3): 233-239.
doi: 10.1038/nbt.2508 pmid: 23360965 |
| [50] |
Lin YN, Cradick TJ, Brown MT, et al. CRISPR/Cas9 systems have off-target activity with insertions or deletions between target DNA and guide RNA sequences[J]. Nucleic Acids Res, 2014, 42(11): 7473-7485.
doi: 10.1093/nar/gku402 pmid: 24838573 |
| [51] |
Doench JG, Fusi N, Sullender M, et al. Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9[J]. Nat Biotechnol, 2016, 34(2): 184-191.
doi: 10.1038/nbt.3437 pmid: 26780180 |
| [52] |
Braglia P, Percudani R, Dieci G. Sequence context effects on oligo(dt)termination signal recognition by Saccharomyces cerevisiae RNA polymerase III*[J]. Journal of Biological Chemistry, 2005, 280(20):19551-19562.
doi: 10.1074/jbc.M412238200 URL |
| [53] |
Langmead B, Trapnell C, Pop M, et al. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome[J]. Genome Biol, 2009, 10(3): R25.
doi: 10.1186/gb-2009-10-3-r25 URL |
| [54] |
Ronda C, Pedersen LE, Hansen HG, et al. Accelerating genome editing in CHO cells using CRISPR Cas9 and CRISPy, a web-based target finding tool[J]. Biotechnol Bioeng, 2014, 111(8): 1604-1616.
doi: 10.1002/bit.25233 pmid: 24827782 |
| [55] |
Blin K, Pedersen LE, Weber T, et al. CRISPy-web: an online resource to design sgRNAs for CRISPR applications[J]. Synth Syst Biotechnol, 2016, 1(2): 118-121.
doi: 10.1016/j.synbio.2016.01.003 pmid: 29062934 |
| [56] |
Heigwer F, Kerr G, Boutros M. E-CRISP: fast CRISPR target site identification[J]. Nat Methods, 2014, 11(2): 122-123.
doi: 10.1038/nmeth.2812 pmid: 24481216 |
| [57] |
Montague TG, Cruz JM, Gagnon JA, et al. CHOPCHOP: a CRISPR/Cas9 and TALEN web tool for genome editing[J]. Nucleic Acids Res, 2014, 42(W1): W401-W407.
doi: 10.1093/nar/gku410 URL |
| [58] |
Horwitz AA, Walter JM, Schubert MG, et al. Efficient multiplexed integration of synergistic alleles and metabolic pathways in yeasts via CRISPR-cas[J]. Cell Syst, 2015, 1(1): 88-96.
doi: 10.1016/j.cels.2015.02.001 pmid: 27135688 |
| [59] |
Bao ZH, Xiao H, Liang J, et al. Homology-integrated CRISPR-cas(HI-CRISPR)system for one-step multigene disruption in Saccharomyces cerevisiae[J]. ACS Synth Biol, 2015, 4(5): 585-594.
doi: 10.1021/sb500255k URL |
| [60] |
Ferreira R, Skrekas C, Nielsen J, et al. Multiplexed CRISPR/Cas9 genome editing and gene regulation using Csy4 in Saccharomyces cerevisiae[J]. ACS Synth Biol, 2018, 7(1): 10-15.
doi: 10.1021/acssynbio.7b00259 pmid: 29161506 |
| [61] |
Hopper AK. Transfer RNA post-transcriptional processing, turnover, and subcellular dynamics in the yeast Saccharomyces cerevisiae[J]. Genetics, 2013, 194(1): 43-67.
doi: 10.1534/genetics.112.147470 pmid: 23633143 |
| [62] |
Papadimitriou A, Gross HJ. Pre-tRNA 3'-processing in Saccharo-myces cerevisiae. purification and characterization of exo-and endoribonucleases[J]. Eur J Biochem, 1996, 242(3): 747-759.
pmid: 9022706 |
| [63] |
Lan PF, Tan M, Zhang YB, et al. Structural insight into precursor tRNA processing by yeast ribonuclease P[J]. Science, 2018, 362(6415): eaat6678.
doi: 10.1126/science.aat6678 URL |
| [64] |
Zhang YP, Wang J, Wang ZB, et al. A gRNA-tRNA array for CRISPR-Cas9 based rapid multiplexed genome editing in Saccharomyces cerevisiae[J]. Nat Commun, 2019, 10(1): 1053.
doi: 10.1038/s41467-019-09005-3 |
| [65] |
Ziehler WA, Day JJ, Fierke CA, et al. Effects of 5' leader and 3' trailer structures on pre-tRNA processing by nuclear RNase P[J]. Biochemistry, 2000, 39(32): 9909-9916.
pmid: 10933810 |
| [66] |
Gao YB, Zhao YD. Self-processing of ribozyme-flanked RNAs into guide RNAs in vitro and in vivo for CRISPR-mediated genome editing[J]. J Integr Plant Biol, 2014, 56(4): 343-349.
doi: 10.1111/jipb.v56.4 URL |
| [67] |
Ryan OW, Skerker JM, Maurer MJ, et al. Selection of chromosomal DNA libraries using a multiplex CRISPR system[J]. eLife, 2014, 3: e03703.
doi: 10.7554/eLife.03703 URL |
| [68] |
DiCarlo JE, Chavez A, Dietz SL, et al. Safeguarding CRISPR-Cas9 gene drives in yeast[J]. Nat Biotechnol, 2015, 33(12): 1250-1255.
doi: 10.1038/nbt.3412 pmid: 26571100 |
| [69] | Zhang F. CRISPR-Cas systems and methods for altering expression of gene products:US8697359(B1)[P].2016-10-28 |
| [70] |
Pliatsika V, Rigoutsos I. “Off-Spotter”: very fast and exhaustive enumeration of genomic lookalikes for designing CRISPR/Cas guide RNAs[J]. Biol Direct, 2015, 10(1): 1-10.
doi: 10.1186/s13062-014-0031-8 URL |
| [71] | 陈小玲, 黄尚飞. 一种快速检查基因打靶特异性的方法及系统: 中国, 202310802061.9[P]. 2023-07-03. |
| Chen XL, Huang SF. A method and system for rapid detection of DNA targeting specificity: CN202310802061.9[P]. 2023-07-03. | |
| [72] | 陈小玲, 黄尚飞, 陈英, 等. 一种设计靶序列的方法及系统: CN202310802050.0[P].2023-07-03. |
| Chen XL, Huang SF, Chen Y, et al. A method and system for designing target sequences: CN202310802050.0[P]. 2023-07-03. |
| [1] | ZHANG Chen, LEI Zhan, LI Kai, SHANG Ying, XU Wen-tao. Research Progress on Off-target Effects and Detection Techniques in CRISPR/Cas9 Systems [J]. Biotechnology Bulletin, 2020, 36(3): 78-87. |
| [2] | WU Yan, HAO Ya-qiao, WEI Xuan, SHEN Qi, LIU Ye-fei, WANG Sheng-hou, ZHAO Hong-xin. The Advantages and Limitations of CRISPR/Cas9-based Gene Editing Technology [J]. Biotechnology Bulletin, 2018, 34(5): 1-8. |
| [3] | BI Yan-zhen, XIAO Hong-wei, ZHANG Li-ping, REN Hong-yan, HUA Zai-dong, HUA Wen-jun, WANG Zheng, NIU Min-jie, LIN Zheng-yun, REN Xi-dong, SUN Li-hua, ZHENG Xin-min. Applications and Challenges of Gene Editing in Non-human Primate Models [J]. Biotechnology Bulletin, 2018, 34(5): 48-56. |
| [4] | DONG Wei-peng, WANG Jun-shi, ZHANG Shao-hua, YAN Jiong. Research Progress of CRISPR System and Its Application in Mice [J]. Biotechnology Bulletin, 2018, 34(5): 57-63. |
| [5] | YUAN Wei-xi, YU Yun-mei, HU Chun-cai, ZHAO Zu-guo. Current Issues and Progress in the Application of CRISPR/Cas9 Technique [J]. Biotechnology Bulletin, 2017, 33(4): 70-77. |
| [6] | YIN Shen, HE Gui-fang, LAI Fang-nong, XIE Feng-yun, MA Jun-yu. Off-target of CRISPR/Cas9 System [J]. Biotechnology Bulletin, 2016, 32(3): 31-37. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||