








Biotechnology Bulletin ›› 2023, Vol. 39 ›› Issue (8): 241-250.doi: 10.13560/j.cnki.biotech.bull.1985.2022-1562
Previous Articles Next Articles
LI Bo1,3( ), LIU He-xia1, CHEN Yu-ling1, ZHOU Xing-wen2, ZHU Yu-lin1,3(
), LIU He-xia1, CHEN Yu-ling1, ZHOU Xing-wen2, ZHU Yu-lin1,3( )
)
Received:2022-12-29
Online:2023-08-26
Published:2023-09-05
Contact:
ZHU Yu-lin
E-mail:lbshaojianbo@ylu.edu.cn;gxzyl@163.com
LI Bo, LIU He-xia, CHEN Yu-ling, ZHOU Xing-wen, ZHU Yu-lin. Cloning, Subcellular Localization and Expression Analysis of CnbHLH79 Transcription Factor from Camellia nitidissima[J]. Biotechnology Bulletin, 2023, 39(8): 241-250.

Fig. 1 Different cultures of C. nitidissima for fluorescence quantitative analysis The cultures of C. nitidissima are lateral root, stem and leave from left to right. The different flowering stages are young bud(A), early bud(B), chromogenic stages of flowering(C), the half-opening stage(D)and blooming stage(E)
| 引物名称 Primer name | 引物序列 Primer sequence(5'-3') | 用途 Use |
|---|---|---|
| CnbHLH79-F | GATCCTCCGATAATCAATTTGACCTCTTTC | 克隆Clone |
| CnbHLH79-R | TGCTGCTCTATCAAAACTGCTACCAAC | |
| CnbHLH79-F | GGAACACCAACATTTGATGC | 定量表达Quantitative expression |
| CnbHLH79-R | ATGAAGCCATTCGGTTTGT | |
| CnbHLH79-F | ACTAGGGTCTCGCACCATGGATCCTCCGATAATCAATTTGACCTCTTTC | 亚细胞定位Subcellular localization |
| CnbHLH79-R | ACTAGGGTCTCTCGCC TGCTGCTCTATCAAAACTGCTACCAAC | |
| 18S rRNA-F | CAACCATAAACGATGCCGA | 内参基因Reference gene |
| 18S rRNA-R | AGCCTTGCGACCATACTCC |
Table 1 Primers used for cloning, subcellular localization and quantitative analysis of CnbHLH79
| 引物名称 Primer name | 引物序列 Primer sequence(5'-3') | 用途 Use |
|---|---|---|
| CnbHLH79-F | GATCCTCCGATAATCAATTTGACCTCTTTC | 克隆Clone |
| CnbHLH79-R | TGCTGCTCTATCAAAACTGCTACCAAC | |
| CnbHLH79-F | GGAACACCAACATTTGATGC | 定量表达Quantitative expression |
| CnbHLH79-R | ATGAAGCCATTCGGTTTGT | |
| CnbHLH79-F | ACTAGGGTCTCGCACCATGGATCCTCCGATAATCAATTTGACCTCTTTC | 亚细胞定位Subcellular localization |
| CnbHLH79-R | ACTAGGGTCTCTCGCC TGCTGCTCTATCAAAACTGCTACCAAC | |
| 18S rRNA-F | CAACCATAAACGATGCCGA | 内参基因Reference gene |
| 18S rRNA-R | AGCCTTGCGACCATACTCC |

Fig. 3 Prediting functional domain of protein CnbHLH79 and amino acid multiple sequence aligning in C. nitidissima A: Protein functional domains prediction(bHLH_AtBPE_like: conserved domains; bHLH_SF: superfamily). B: Homology comparison of putative amino acids sequence. The sequence underlined in red: conserved domains of bHLH_SF superfamily; conserved amino acids of bHLH_SF superfamily contained by bHLH79 protein; α1 and α2: α-helix; CnbHLH79: bHLH79 transcription factor of C. nitidissima; AtbHLH79: bHLH79 transcription factor of Arabidopsis thaliana(AT5G62610.1); CsbHLH79_like: bHLH79 transcription factor of C. sinensis(XP_028119408.1); ArBPEp: BPEp transcription factor of A. rufa(GFS32687.1)
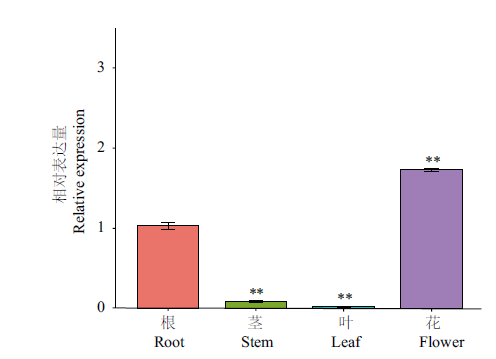
Fig. 7 Relative expressions of CnbHLH79 in different culture ** indicates extremely significant difference in the expression of stem, leave and flower compared with root(P<0.01). Reference gene: 18S rRNA; n=3
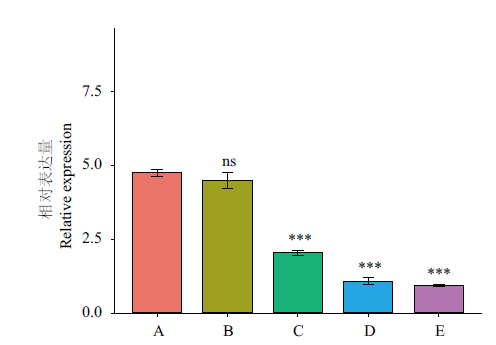
Fig. 8 Expressions of CnbHLH79 at different flowering stages A: Young bud; B: early bud; C: chromogenic stages of flowering; D: the half-opening stage; E: blooming stage. *** indicates the expressions of C, D, and E shows extremely significant difference compared with that of A(P<0.001). ns means no significant difference. Reference gene: 18S rRNA; n=3
| [1] | 柴胜丰, 韦霄, 蒋运生, 等. 濒危植物金花茶开花物候和生殖构件特征[J]. 热带亚热带植物学报, 2009, 17(1): 5-11. |
| Chai SF, Wei X, Jiang YS, et al. The flowering phenology and characteristics of reproductive modules of endangered plant Camellia nitidissima[J]. J Trop Subtrop Bot, 2009, 17(1): 5-11. | |
| [2] | 陈银霞, 唐山, 赵松子. 金花茶花色遗传研究进展[J]. 南方林业科学, 2015, 43(6): 39-41, 55. |
| Chen YX, Tang S, Zhao SZ. Recent advances in flower color genetics of Camellia nitidissima[J]. South China For Sci, 2015, 43(6): 39-41, 55. | |
| [3] |
Peng X, Yu DY, Feng BM, et al. A new acylated flavonoid glycoside from the flowers of Camellia nitidissima and its effect on the induction of apoptosis in human lymphoma U937 cells[J]. J Asian Nat Prod Res, 2012, 14(8): 799-804.
doi: 10.1080/10286020.2012.691475 URL |
| [4] |
Zhou XW, Li JY, Zhu YL, et al. De novo assembly of the Camellia nitidissima transcriptome reveals key genes of flower pigment biosynthesis[J]. Front Plant Sci, 2017, 8: 1545.
doi: 10.3389/fpls.2017.01545 URL |
| [5] |
Morita Y, Hoshino A. Recent advances in flower color variation and patterning of Japanese morning glory and petunia[J]. Breed Sci, 2018, 68(1): 128-138.
doi: 10.1270/jsbbs.17107 URL |
| [6] |
Wu MJ, Lyu XL, Zhou YJ, et al. High anthocyanin accumulation in an Arabidopsis mutant defective in chloroplast biogenesis[J]. Plant Growth Regul, 2019, 87(3): 433-444.
doi: 10.1007/s10725-019-00481-7 |
| [7] |
Jiang LN, Fan ZQ, Tong R, et al. Functional diversification of the dihydroflavonol 4-reductase from Camellia nitidissima Chi. in the control of polyphenol biosynthesis[J]. Genes, 2020, 11(11): 1341.
doi: 10.3390/genes11111341 URL |
| [8] |
Jiang LN, Fan ZQ, Tong R, et al. Flavonoid 3'-hydroxylase of Camellia nitidissima Chi. promotes the synthesis of polyphenols better than flavonoids[J]. Mol Biol Rep, 2021, 48(5): 3903-3912.
doi: 10.1007/s11033-021-06345-6 |
| [9] |
Zhou XW, Fan ZQ, Chen Y, et al. Functional analyses of a flavonol synthase-like gene from Camellia nitidissima reveal its roles in flavonoid metabolism during floral pigmentation[J]. J Biosci, 2013, 38(3): 593-604.
doi: 10.1007/s12038-013-9339-2 URL |
| [10] | 周兴文, 李纪元, 范正琪. 金花茶查尔酮合成酶基因全长克隆与序列分析[J]. 生物技术通报, 2011(6): 58-64. |
| Zhou XW, Li JY, Fan ZQ. Cloning and sequence analysis of Chalcone synthase gene cDNA from Camellia nitidissima[J]. Biotechnol Bull, 2011(6): 58-64. | |
| [11] |
周兴文, 李纪元, 朱宇林. 金花茶查尔酮合成酶基因CnCHS的克隆及遗传转化研究[J]. 植物研究, 2015, 35(3): 327-332.
doi: 10.7525/j.issn.1673-5102.2015.03.002 |
| Zhou XW, Li JY, Zhu YL. Cloning and genetic transformation of CnCHS gene from Camellia nitidissima[J]. Bull Bot Res, 2015, 35(3): 327-332. | |
| [12] |
Xu WJ, Dubos C, Lepiniec L. Transcriptional control of flavonoid biosynthesis by MYB-bHLH-WDR complexes[J]. Trends Plant Sci, 2015, 20(3): 176-185.
doi: 10.1016/j.tplants.2014.12.001 pmid: 25577424 |
| [13] |
Zhao L, Song ZB, Wang BW, et al. R2R3-MYB transcription factor NtMYB330 regulates proanthocyanidin biosynthesis and seed germination in tobacco(Nicotiana tabacum L.)[J]. Front Plant Sci, 2022, 12: 819247.
doi: 10.3389/fpls.2021.819247 URL |
| [14] |
Zhong CM, Tang Y, Pang B, et al. The R2R3-MYB transcription factor GhMYB1a regulates flavonol and anthocyanin accumulation in Gerbera hybrida[J]. Hortic Res, 2020, 7: 78.
doi: 10.1038/s41438-020-0296-2 |
| [15] |
Sun BM, Zhu ZS, Cao PR, et al. Purple foliage coloration in tea(Camellia sinensis L.) arises from activation of the R2R3-MYB transcription factor CsAN1[J]. Sci Rep, 2016, 6: 32534.
doi: 10.1038/srep32534 |
| [16] |
Albert NW, Davies KM, Lewis DH, et al. A conserved network of transcriptional activators and repressors regulates anthocyanin pigmentation in eudicots[J]. Plant Cell, 2014, 26(3): 962-980.
doi: 10.1105/tpc.113.122069 URL |
| [17] |
Zhao AJ, Cui Z, Li TG, et al. mRNA and miRNA expression analysis reveal the regulation for flower spot patterning in Phalaenopsis ‘Panda’[J]. Int J Mol Sci, 2019, 20(17): 4250.
doi: 10.3390/ijms20174250 URL |
| [18] |
Yamagishi M. Isolation and identification of MYB transcription factors(MYB19Long and MYB19Short)involved in raised spot anthocyanin pigmentation in lilies(Lilium spp.)[J]. J Plant Physiol, 2020, 250: 153164.
doi: 10.1016/j.jplph.2020.153164 URL |
| [19] |
Li YQ, Shan XT, Tong LN, et al. The conserved and particular roles of the R2R3-MYB regulator FhPAP1 from Freesia hybrida in flower anthocyanin biosynthesis[J]. Plant Cell Physiol, 2020, 61(7): 1365-1380.
doi: 10.1093/pcp/pcaa065 URL |
| [20] |
郑清冬, 王艺, 欧悦, 等. 兰科植物花色相关基因研究进展[J]. 园艺学报, 2021, 48(10): 2057-2072.
doi: 10.16420/j.issn.0513-353x.2021-0444 |
|
Zheng QD, Wang Y, Ou Y, et al. Research advances of genes responsible for flower colors in Orchidaceae[J]. Acta Hortic Sin, 2021, 48(10): 2057-2072.
doi: 10.16420/j.issn.0513-353x.2021-0444 |
|
| [21] | 王玉书, 杨旭妍, 付震, 等. 观赏羽衣甘蓝BoDFR基因的克隆及表达分析[J]. 西北植物学报, 2020, 40(9): 1483-1489. |
| Wang YS, Yang XY, Fu Z, et al. Cloning and expression analysis of DFR in ornamental kale(Brassica oleracea L. var. acephala)[J]. Acta Bot Boreali Occidentalia Sin, 2020, 40(9): 1483-1489. | |
| [22] |
Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method[J]. Methods, 2001, 25(4): 402-408.
doi: 10.1006/meth.2001.1262 pmid: 11846609 |
| [23] |
Feller A, Machemer K, Braun EL, et al. Evolutionary and comparative analysis of MYB and bHLH plant transcription factors[J]. Plant J, 2011, 66(1): 94-116.
doi: 10.1111/tpj.2011.66.issue-1 URL |
| [24] |
Hao YQ, Zong XM, Ren P, et al. Basic helix-loop-helix(bHLH)transcription factors regulate a wide range of functions in Arabidopsis[J]. Int J Mol Sci, 2021, 22(13): 7152.
doi: 10.3390/ijms22137152 URL |
| [25] |
Matus JT, Poupin MJ, Cañón P, et al. Isolation of WDR and bHLH genes related to flavonoid synthesis in grapevine(Vitis vinifera L.)[J]. Plant Mol Biol, 2010, 72(6): 607-620.
doi: 10.1007/s11103-010-9597-4 pmid: 20112051 |
| [26] |
Kim DH, Park S, Lee JY, et al. Enhancing flower color through simultaneous expression of the B-Peru and mPAP1 transcription factors under control of a flower-specific promoter[J]. Int J Mol Sci, 2018, 19(1): 309.
doi: 10.3390/ijms19010309 URL |
| [27] | Deng CY, Wang JY, Lu CF, et al. CcMYB6-1 and CcbHLH1, two novel transcription factors synergistically involved in regulating anthocyanin biosynthesis in cornflower[J]. Plant Physiol Biochem, 2020, 151: 271-283. |
| [28] |
Li CH, Qiu J, Ding L, et al. Anthocyanin biosynthesis regulation of DhMYB2 and DhbHLH1 in Dendrobium hybrids petals[J]. Plant Physiol Biochem, 2017, 112: 335-345.
doi: 10.1016/j.plaphy.2017.01.019 URL |
| [29] |
Liu HX, Liu Q, Chen YL, et al. Full-length transcriptome sequencing provides insights into flavonoid biosynthesis in Camellia nitidissima Petals[J]. Gene, 2023, 850: 146924.
doi: 10.1016/j.gene.2022.146924 URL |
| [30] | 李辛雷, 王佳童, 孙振元, 等. 五种金花茶组植物类黄酮成分及其与花色关系[J]. 生态学杂志, 2019, 38(4): 961-966. |
| Li XL, Wang JT, Sun ZY, et al. Flavonoid components and their relationship with flower colors in five species of Camellia section Chrysantha[J]. Chin J Ecol, 2019, 38(4): 961-966. | |
| [31] | 姜丽娜, 李纪元, 童冉, 等. 金花茶组植物花色与细胞内重要环境因子的关系[J]. 广西植物, 2019, 39(12): 1605-1612. |
| Jiang LN, Li JY, Tong R, et al. Relationship between flower color and important cellular environment elemental factors in yellow Camellia[J]. Guihaia, 2019, 39(12): 1605-1612. | |
| [32] |
Winkel-Shirley B. Flavonoid biosynthesis. A colorful model for genetics, biochemistry, cell biology, and biotechnology[J]. Plant Physiol, 2001, 126(2): 485-493.
doi: 10.1104/pp.126.2.485 pmid: 11402179 |
| [33] |
Agati G, Tattini M. Multiple functional roles of flavonoids in photoprotection[J]. New Phytol, 2010, 186(4): 786-793.
doi: 10.1111/j.1469-8137.2010.03269.x pmid: 20569414 |
| [34] | Jaakola L, Hohtola A. Effect of latitude on flavonoid biosynthesis in plants[J]. Plant Cell Environ, 2010, 33(8): 1239-1247. |
| [35] |
An JP, Li R, Qu FJ, et al. R2R3-MYB transcription factor MdMYB23 is involved in the cold tolerance and proanthocyanidin accumulation in apple[J]. Plant J, 2018, 96(3): 562-577.
doi: 10.1111/tpj.14050 URL |
| [36] |
Samarina LS, Bobrovskikh AV, Doroshkov AV, et al. Comparative expression analysis of stress-inducible candidate genes in response to cold and drought in tea plant[Camellia sinensis(L.) kuntze][J]. Front Genet, 2020, 11: 611283.
doi: 10.3389/fgene.2020.611283 URL |
| [37] |
Wang ZY, Zhang YM, Hu HF, et al. CabHLH79 acts upstream of CaNAC035 to regulate cold stress in pepper[J]. Int J Mol Sci, 2022, 23(5): 2537.
doi: 10.3390/ijms23052537 URL |
| [1] | YANG Zhi-xiao, HOU Qian, LIU Guo-quan, LU Zhi-gang, CAO Yi, GOU Jian-yu, WANG Yi, LIN Ying-chao. Responses of Rubisco and Rubisco Activase in Different Resistant Tobacco Strains to Brown Spot Stress [J]. Biotechnology Bulletin, 2023, 39(9): 202-212. |
| [2] | LYU Qiu-yu, SUN Pei-yuan, RAN Bin, WANG Jia-rui, CHEN Qing-fu, LI Hong-you. Cloning, Subcellular Localization and Expression Analysis of the Transcription Factor Gene FtbHLH3 in Fagopyrum tataricum [J]. Biotechnology Bulletin, 2023, 39(8): 194-203. |
| [3] | YANG Xu-yan, ZHAO Shuang, MA Tian-yi, BAI Yu, WANG Yu-shu. Cloning of Three Cabbage WRKY Genes and Their Expressions in Response to Abiotic Stress [J]. Biotechnology Bulletin, 2023, 39(11): 261-269. |
| [4] | GUO Zhi-hao, JIN Ze-xin, LIU Qi, GAO Li. Bioinformatics Analysis, Subcellular Localization and Toxicity Verification of Effector g11335 in Tilletia contraversa Kühn [J]. Biotechnology Bulletin, 2022, 38(8): 110-117. |
| [5] | YANG Jia-bao, ZHOU Zhi-ming, ZHANG Zhan, FENG Li, SUN Li. Cloning,Expression of Helianthus annuus HaLACS1 Gene and Identification of Its Functional Complementation in Saccharomyces cerevisiae [J]. Biotechnology Bulletin, 2022, 38(6): 147-156. |
| [6] | YU Guo-hong, LIU Peng-cheng, LI Lei, LI Ming-zhe, CUI Hai-ying, HAO Hong-bo, GUO An-qiang. Physiological Responses of Potato in Different Genotypes to Drought Stress [J]. Biotechnology Bulletin, 2022, 38(5): 56-63. |
| [7] | HAO Qing-qing, YAO Sheng, LIU Jia-he, CHEN Pei-zhen, ZHANG Meng-yang, JI Kong-shu. Cloning and Expression Analysis of NAC Transcription Factor PmNAC8 in Pinus massoniana [J]. Biotechnology Bulletin, 2022, 38(4): 202-216. |
| [8] | DANG Yuan, LI Wei, MIAO Xiang, XIU Yu, LIN Shan-zhi. Cloning of Oleosin Gene PsOLE4 from Prunus sibirica and Its Regulatory Function Analysis for Oil Accumulation [J]. Biotechnology Bulletin, 2022, 38(11): 151-161. |
| [9] | LUO Ying, TANG Zhi, WANG Fan, LIU Xiao-xia, LUO Xiao-fang, HE Fu-lin. Cloning and Response Analysis to Abiotic Stress of GbR2R3-MYB1 Gene from Ginkgo biloba [J]. Biotechnology Bulletin, 2022, 38(10): 184-194. |
| [10] | SUN Rui-fen, ZHANG Yan-fang, NIU Su-qing, GUO Shu-chun, LI Su-ping, YU Hai-feng, NIE Hui, MOU Ying-nan. Expression Analysis and Functional Verification of the HaACO1 Gene in Sunflower [J]. Biotechnology Bulletin, 2021, 37(9): 114-124. |
| [11] | FAN Ya-peng, RUI Cun, ZHANG Yue-xin, CHEN Xiu-gui, LU Xu-ke, WANG Shuai, ZHANG Hong, XU Nan, WANG Jing, CHEN Chao, YE Wu-wei. Cloning,Expression and Preliminary Bioinformatics Analysis of the Alkaline Tolerant Gene GhZAT12 in Gossypium hirsutum [J]. Biotechnology Bulletin, 2021, 37(8): 121-130. |
| [12] | SHAN Cao-mei, YE Lei, ZHANG Lian-hu, KUANG Wei-gang, SUN Xiao-tang, MA Jian, CUI Ru-qiang. Cloning,and Functional Analysis of Gene OsRAI1 Resistant to Hirschmanniella mucronate in Rice [J]. Biotechnology Bulletin, 2021, 37(7): 146-155. |
| [13] | SUN Xiao-qian, WANG Jia-rui, CHEN Qing-fu, LI Hong-you. Gene Cloning,Subcellular Localization and Expression Analyses of FtMYBF Transcription Factor in Fagopyrum tataricum [J]. Biotechnology Bulletin, 2021, 37(3): 10-17. |
| [14] | HAN Zhan-hong, ZONG Yuan-yuan, ZHANG Xue-mei, WANG Bin, PRUSKY Dov, BI Yang. Bioinformatics,Subcellular Localization and Expression Analysis of erg4 in Penicillium expansum [J]. Biotechnology Bulletin, 2021, 37(12): 60-70. |
| [15] | ZHANG Rui-zhu, JIANG Yu-chen, HUANG Jun, YAN Jie. Cloning and Expression Analysis of SRPP2 Gene in Taraxacum kok-saghyz Rodin [J]. Biotechnology Bulletin, 2020, 36(1): 9-14. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||