








Biotechnology Bulletin ›› 2023, Vol. 39 ›› Issue (8): 251-261.doi: 10.13560/j.cnki.biotech.bull.1985.2022-1557
Previous Articles Next Articles
FU Yu( ), JIA Rui-rui, HE He, WANG Liang-gui, YANG Xiu-lian(
), JIA Rui-rui, HE He, WANG Liang-gui, YANG Xiu-lian( )
)
Received:2022-12-27
Online:2023-08-26
Published:2023-09-05
Contact:
YANG Xiu-lian
E-mail:1091322721@qq.com;xly@njfu.edu.cn
FU Yu, JIA Rui-rui, HE He, WANG Liang-gui, YANG Xiu-lian. Growth Differences Among Grafted Seedlings with Two Rootstocks of Catalpa bungei and Comparative Analysis of Transcriptome[J]. Biotechnology Bulletin, 2023, 39(8): 251-261.
| 嫁接组合 Grafting combination | 取样部位 Sampling part | 编号 Numbering |
|---|---|---|
| 苏楸1号/梓树 ‘Suqiu No. 1’/ Catalpa ovata | 叶片Leaf | ZS-L |
| 顶芽Bud | ZS-B | |
| 苏楸1号/滇楸 ‘Suqiu No. 1’/Catalpa fargesii | 叶片Leaf | DS-L |
| 顶芽Bud | DS-B |
Table1 Grafting combination and sampling part
| 嫁接组合 Grafting combination | 取样部位 Sampling part | 编号 Numbering |
|---|---|---|
| 苏楸1号/梓树 ‘Suqiu No. 1’/ Catalpa ovata | 叶片Leaf | ZS-L |
| 顶芽Bud | ZS-B | |
| 苏楸1号/滇楸 ‘Suqiu No. 1’/Catalpa fargesii | 叶片Leaf | DS-L |
| 顶芽Bud | DS-B |
| 引物名称Primer name | 序列Sequence(5'-3') |
|---|---|
| CfMADH-F | AGCTTCCATTCTTTGCCTCA |
| CfMADH-R | TCCGAACAAAAGCAATACCC |
| Unigene0036149-F | AGGAACAGGAGAATGTGAAGAGG |
| Unigene0036149-R | CCGTCGTCACTTCTCGCAG |
| Unigene0007805-F | CAAAAAATGCTGGAAAAGTACCC |
| Unigene0007805-R | TGGTAAAATCGTGGTTCTCAAGTG |
| Unigene0040270-F | CCGCTTCATCTTGGTTCAGTG |
| Unigene0040270-R | AAGAAGGAAGATTTGCTGTTGGA |
| Unigene0006320-F | AGAAACACGAAAGTGAATGCTAAAC |
| Unigene0006320-R | GCACTCGGATGAAACGAAAAC |
Table 2 Primer sequences of RT-qPCR
| 引物名称Primer name | 序列Sequence(5'-3') |
|---|---|
| CfMADH-F | AGCTTCCATTCTTTGCCTCA |
| CfMADH-R | TCCGAACAAAAGCAATACCC |
| Unigene0036149-F | AGGAACAGGAGAATGTGAAGAGG |
| Unigene0036149-R | CCGTCGTCACTTCTCGCAG |
| Unigene0007805-F | CAAAAAATGCTGGAAAAGTACCC |
| Unigene0007805-R | TGGTAAAATCGTGGTTCTCAAGTG |
| Unigene0040270-F | CCGCTTCATCTTGGTTCAGTG |
| Unigene0040270-R | AAGAAGGAAGATTTGCTGTTGGA |
| Unigene0006320-F | AGAAACACGAAAGTGAATGCTAAAC |
| Unigene0006320-R | GCACTCGGATGAAACGAAAAC |
| 嫁接组合 Grafting combination | 成活率 Survival rate/% | 新梢长度 New shoot length/cm | 新梢粗度 New shoot thickness/cm | 接穗粗度/砧木粗度 Scion diameter/Stock diameter | 叶面积 Leaf area/cm2 |
|---|---|---|---|---|---|
| ZS | 60.95±0.064** | 107.51±12.25** | 2.47±0.65** | 0.822±0.029** | 338.64±6.11** |
| DS | 32.05±0.038 | 83.43±5.06 | 2.04±1.57 | 0.721±0.087 | 247.61±6.59 |
Table 3 Survival rates and growths of grafted seedlings of two Catalpa trees
| 嫁接组合 Grafting combination | 成活率 Survival rate/% | 新梢长度 New shoot length/cm | 新梢粗度 New shoot thickness/cm | 接穗粗度/砧木粗度 Scion diameter/Stock diameter | 叶面积 Leaf area/cm2 |
|---|---|---|---|---|---|
| ZS | 60.95±0.064** | 107.51±12.25** | 2.47±0.65** | 0.822±0.029** | 338.64±6.11** |
| DS | 32.05±0.038 | 83.43±5.06 | 2.04±1.57 | 0.721±0.087 | 247.61±6.59 |
| 高质量数据碱基总数 Clean data/bp | Q20/% | Q30/% | N/% | GC/% |
|---|---|---|---|---|
| 75 612 886 298 | 96.65-97.36 | 91.16-92.69 | 0 | 43.79-44.57 |
Table 4 Summary results of sequencing data for 12 samples
| 高质量数据碱基总数 Clean data/bp | Q20/% | Q30/% | N/% | GC/% |
|---|---|---|---|---|
| 75 612 886 298 | 96.65-97.36 | 91.16-92.69 | 0 | 43.79-44.57 |
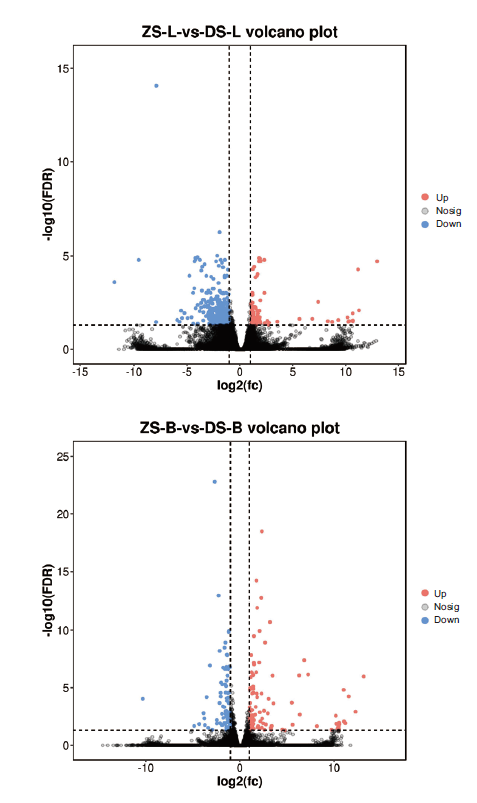
Fig. 2 Volcano plot of different genes in different grafting combinations The abscissa indicates the multiple log2 value of the difference between the two groups, the ordinate indicates the negative Log10 value of the FDR of the difference between the two groups. Red indicates the expression was up-regulated and blue was down-regulated
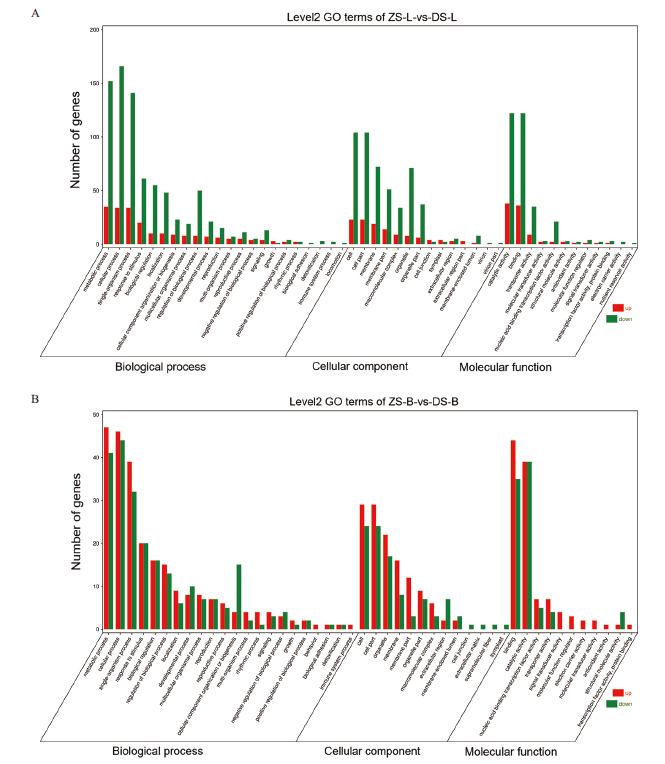
Fig. 3 GO annotation enrichment analysis diagram of differential genes in different grafting combinations A:ZS-LvsDS-L; B:ZS-BvsDS-B. The abscissa represents GO Term level 1 and 2, the ordinate represents the number of differentially expressed genes
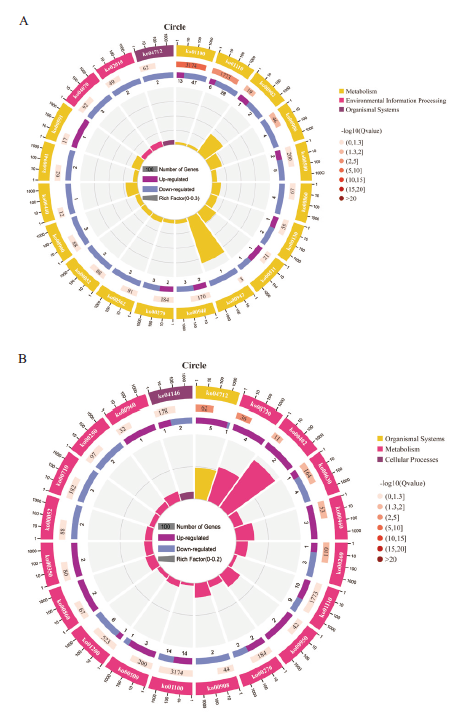
Fig. 4 KEGG annotation enrichment analysis diagram of differential genes in different grafting combinations A:ZS-L vs DS-L;B:ZS-B vs DS-B. The first circle: enrich the top 20 pathways, outside the circle is the coordinate ruler of the number of differential genes. Different colors indicate different A classes. The second circle: the number and Q value of the pathway in the differential gene background. The more the number of differential genes background, the longer the bar, the smaller the Q value, the redder the color. The third circle: the bar graph of the ratio of up- and down-regulated differential genes, dark purple represents the ratio of up-regulated differential genes, and light purple represents the ratio of down-regulated differential genes; the lower part shows the specific value. The fourth circle: the RichFactor value of each pathway(the number of differential genes in the pathway divided by all the numbers in the pathway), background grid lines, each grid indicates 0.1
| 通路Pathway | 基因ID Unigene ID | 基因名称Symbol | 描述[ID]Description[ID] |
|---|---|---|---|
| 类胡萝卜素生物合成 Carotenoid biosynthesis | Unigene0051519 | PSY1 | 顺式八氢番茄红素合酶[EC:2.5.1.32]15-cis-Phytoene synthase |
| Unigene0046587 | LCY1 | 番茄红素β-环化酶[EC:5.5.1.19]Lycopene beta-cyclase | |
| Unigene0013377 | NCED2 | 9-顺式环氧类胡萝卜素双加氧酶[EC:1.13.11.51] 9-cis-Epoxycarotenoid dioxygenase | |
| Unigene0044139 | CYP707A2 | 脱落酸-8'-羟化酶[EC:1.14.14.137]Abscisic acid 8'-hydroxylase | |
| 卟啉和叶绿素代谢 Porphyrin and chlorophyll metabolism | Unigene0054531 | HEMA1 | 谷氨酰-tRNA还原酶[EC:1.2.1.70]Glutamyl-tRNA reductase |
| Unigene0017692 | CRD1 | 镁-原卟啉IX单甲酯(氧化)环化[EC:1.14.13.81] Magnesium-protoporphyrin IX monomethyl ester(oxidative)cyclase | |
| Unigene0005359 | CAO | 叶绿素a加氧酶[EC:1.14.13.122]Chlorophyllide a oxygenase | |
| Unigene0040270 | CAO | 叶绿素a加氧酶[EC:1.14.13.122]Chlorophyllide a oxygenase |
Tab.5 Statistics of differentially expressed genes related to photosynthesis
| 通路Pathway | 基因ID Unigene ID | 基因名称Symbol | 描述[ID]Description[ID] |
|---|---|---|---|
| 类胡萝卜素生物合成 Carotenoid biosynthesis | Unigene0051519 | PSY1 | 顺式八氢番茄红素合酶[EC:2.5.1.32]15-cis-Phytoene synthase |
| Unigene0046587 | LCY1 | 番茄红素β-环化酶[EC:5.5.1.19]Lycopene beta-cyclase | |
| Unigene0013377 | NCED2 | 9-顺式环氧类胡萝卜素双加氧酶[EC:1.13.11.51] 9-cis-Epoxycarotenoid dioxygenase | |
| Unigene0044139 | CYP707A2 | 脱落酸-8'-羟化酶[EC:1.14.14.137]Abscisic acid 8'-hydroxylase | |
| 卟啉和叶绿素代谢 Porphyrin and chlorophyll metabolism | Unigene0054531 | HEMA1 | 谷氨酰-tRNA还原酶[EC:1.2.1.70]Glutamyl-tRNA reductase |
| Unigene0017692 | CRD1 | 镁-原卟啉IX单甲酯(氧化)环化[EC:1.14.13.81] Magnesium-protoporphyrin IX monomethyl ester(oxidative)cyclase | |
| Unigene0005359 | CAO | 叶绿素a加氧酶[EC:1.14.13.122]Chlorophyllide a oxygenase | |
| Unigene0040270 | CAO | 叶绿素a加氧酶[EC:1.14.13.122]Chlorophyllide a oxygenase |
| 通路Pathway | 基因ID Unigene ID | 基因名称Symbol | 描述[ID]Description[ID] |
|---|---|---|---|
| 淀粉和蔗糖代谢 Starch and sucrose metabolism | Unigene0038167 | SPS2 | 蔗糖-磷酸合酶[EC:2.4.1.14]Sucrose-phosphate synthase |
| Unigene0005917 | SS2 | 淀粉合酶[EC:2.4.1.21]Starch synthase | |
| Unigene0017971 | TPPJ | 海藻糖6-磷酸磷酸酶[EC:3.1.3.12]Trehalose 6-phosphate phosphatase | |
| Unigene0000390 | GLU3 | 内切葡聚糖酶[EC:3.2.1.4]Endoglucanase | |
| Unigene0022047 | GLU1 | β-葡萄糖苷酶[EC:3.2.1.21]beta-Glucosidase | |
| Unigene0048299 | INV*DC4 | β-果糖呋喃糖苷酶[EC:3.2.1.26]beta-Fructofuranosidase | |
| Unigene0006320 | WAXY | 颗粒结合淀粉合酶[EC:2.4.1.242]Granule-bound starch synthase | |
| Unigene0048994 | STP-1 | 糖原磷酸化酶[EC:2.4.1.1]Glycogen phosphorylase | |
| Unigene0047773 | BAM1 | β-淀粉酶[EC:3.2.1.2]beta-Amylase | |
| Unigene0043824 | BACOVA-02659 | β-葡萄糖苷酶[EC:3.2.1.21]beta-Glucosidase |
Tab.6 Statistics of differentially expressed genes related to starch and sucrose metabolism
| 通路Pathway | 基因ID Unigene ID | 基因名称Symbol | 描述[ID]Description[ID] |
|---|---|---|---|
| 淀粉和蔗糖代谢 Starch and sucrose metabolism | Unigene0038167 | SPS2 | 蔗糖-磷酸合酶[EC:2.4.1.14]Sucrose-phosphate synthase |
| Unigene0005917 | SS2 | 淀粉合酶[EC:2.4.1.21]Starch synthase | |
| Unigene0017971 | TPPJ | 海藻糖6-磷酸磷酸酶[EC:3.1.3.12]Trehalose 6-phosphate phosphatase | |
| Unigene0000390 | GLU3 | 内切葡聚糖酶[EC:3.2.1.4]Endoglucanase | |
| Unigene0022047 | GLU1 | β-葡萄糖苷酶[EC:3.2.1.21]beta-Glucosidase | |
| Unigene0048299 | INV*DC4 | β-果糖呋喃糖苷酶[EC:3.2.1.26]beta-Fructofuranosidase | |
| Unigene0006320 | WAXY | 颗粒结合淀粉合酶[EC:2.4.1.242]Granule-bound starch synthase | |
| Unigene0048994 | STP-1 | 糖原磷酸化酶[EC:2.4.1.1]Glycogen phosphorylase | |
| Unigene0047773 | BAM1 | β-淀粉酶[EC:3.2.1.2]beta-Amylase | |
| Unigene0043824 | BACOVA-02659 | β-葡萄糖苷酶[EC:3.2.1.21]beta-Glucosidase |
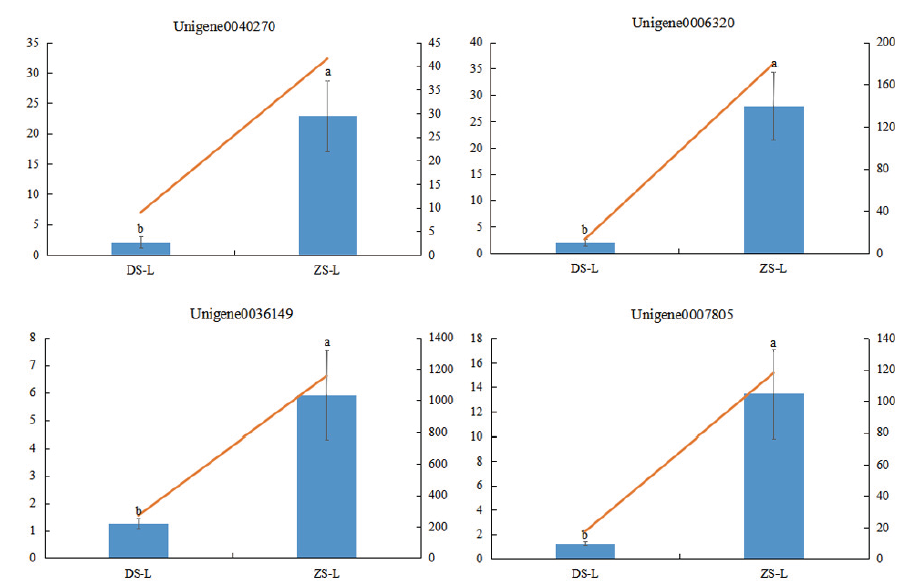
Fig. 7 Relative expressions of four genes in two grafting combinations Different letters indicate the significant difference of relative expression at different grafting combinations.(P<0. 05)
| [1] |
Liu YM, Zhou L, Zhu YQ, et al. Anatomical features and its radial variations among different Catalpa bungei clones[J]. Forests, 2020, 11(8): 824.
doi: 10.3390/f11080824 URL |
| [2] |
Lu N, Zhang MM, Xiao Y, et al. Construction of a high-density genetic map and QTL mapping of leaf traits and plant growth in an interspecific F1 population of Catalpa bungei × Catalpa duclouxii Dode[J]. BMC Plant Biol, 2019, 19(1): 596.
doi: 10.1186/s12870-019-2207-y |
| [3] |
Wu JW, Li JY, Su Y, et al. A morphophysiological analysis of the effects of drought and shade on Catalpa bungei plantlets[J]. Acta Physiol Plant, 2017, 39(3): 80.
doi: 10.1007/s11738-017-2380-2 URL |
| [4] |
Xiao Y, Wang JH, Yun HL, et al. Genetic evaluation and combined selection for the simultaneous improvement of growth and wood properties in Catalpa bungei clones[J]. Forests, 2021, 12(7): 868.
doi: 10.3390/f12070868 URL |
| [5] | 王改萍, 王良桂, 王晓聪, 等. 楸树嫩枝扦插生根发育及根系特征分析[J]. 南京林业大学学报: 自然科学版, 2020, 44(6): 94-102. |
| Wang GP, Wang LG, Wang XC, et al. Dynamic characteristics of cutting rooting of Catalpa bungei with tender branches[J]. J Nanjing For Univ Nat Sci Ed, 2020, 44(6): 94-102. | |
| [6] |
Chen W, Meng PP, Feng H, et al. Effects of arbuscular mycorrhizal fungi on growth and physiological performance of Catalpa bungei C.A.Mey. under drought stress[J]. Forests, 2020, 11(10): 1117.
doi: 10.3390/f11101117 URL |
| [7] | Lin J, Wu LH, Jing L, et al. Effect of different plant growth regulators on callus induction in Catalpa bungei[J]. Afr J Agric Res, 2010, 5: 2699-2704. |
| [8] |
Miao L, Li Q, Sun TS, et al. Sugars promote graft union development in the heterograft of cucumber onto pumpkin[J]. Hortic Res, 2021, 8(1): 146.
doi: 10.1038/s41438-021-00580-5 |
| [9] |
Ren Y, Xu Q, Wang LW, et al. Involvement of metabolic, physiological and hormonal responses in the graft-compatible process of cucumber/pumpkin combinations was revealed through the integrative analysis of mRNA and miRNA expression[J]. Plant Physiol Biochem, 2018, 129: 368-380.
doi: 10.1016/j.plaphy.2018.06.021 URL |
| [10] | Karaca F, Yetİşİr H, Solmaz İ, et al. Rootstock potential of Turkish Lagenaria siceraria germplasm for watermelon: plant growth, yield and quality[J]. Turkish J Agric For, 2012: 167-177. |
| [11] |
Rasool A, Mansoor S, Bhat KM, et al. Mechanisms underlying graft union formation and rootstock scion interaction in horticultural plants[J]. Front Plant Sci, 2020, 11: 590847.
doi: 10.3389/fpls.2020.590847 URL |
| [12] |
Świerczyński S. The effect of rootstock activity for growth and root system soaking in Trichoderma atroviride on the graft success and continued growth of beech(Fagus sylvatica L.) plants[J]. Agronomy, 2022, 12(6): 1259.
doi: 10.3390/agronomy12061259 URL |
| [13] |
Liu WQ, Xiang CG, Li XJ, et al. Identification of long-distance transmissible mRNA between scion and rootstock in cucurbit seedling heterografts[J]. Int J Mol Sci, 2020, 21(15): 5253.
doi: 10.3390/ijms21155253 URL |
| [14] |
Kaseb MO, Umer MJ, Anees M, et al. Transcriptome profiling to dissect the role of genome duplication on graft compatibility mechanisms in watermelon[J]. Biology, 2022, 11(4): 575.
doi: 10.3390/biology11040575 URL |
| [15] |
Liu XY, Li J, Liu MM, et al. Transcriptome profiling to understand the effect of Citrus rootstocks on the growth of ‘shatangju’ mandarin[J]. PLoS One, 2017, 12(1): e0169897.
doi: 10.1371/journal.pone.0169897 URL |
| [16] |
Davoudi M, Song MF, Zhang MR, et al. Long-distance control of the scion by the rootstock under drought stress as revealed by transcriptome sequencing and mobile mRNA identification[J]. Hortic Res, 2022, 9: uhab033.
doi: 10.1093/hr/uhab033 URL |
| [17] | 申妍颖, 李雪涵, 李飞鸿, 等. 中间砧‘南通小方柿’嫁接柿转录组测序及分析[J]. 分子植物育种, 2019, 17(5): 1454-1466. |
| Shen YY, Li XH, Li FH, et al. Transcriptome sequencing and analysis of grafted persimmon with ‘Nantong-Xiaofangshi’ as the interstock[J]. Mol Plant Breed, 2019, 17(5): 1454-1466. | |
| [18] |
Li GF, Ma JJ, Tan M, et al. Transcriptome analysis reveals the effects of sugar metabolism and auxin and cytokinin signaling pathways on root growth and development of grafted apple[J]. BMC Genom, 2016, 17(1): 150.
doi: 10.1186/s12864-016-2484-x URL |
| [19] |
Young MD, Wakefield MJ, Smyth GK, et al. Gene ontology analysis for RNA-seq: accounting for selection bias[J]. Genome Biol, 2010, 11(2): R14.
doi: 10.1186/gb-2010-11-2-r14 URL |
| [20] |
Mao XZ, Cai T, Olyarchuk JG, et al. Automated genome annotation and pathway identification using the KEGG Orthology(KO)as a controlled vocabulary[J]. Bioinformatics, 2005, 21(19): 3787-3793.
doi: 10.1093/bioinformatics/bti430 URL |
| [21] | 杨英英, 赵林姣, 杨桂娟, 等. ‘麦缘锦楸’叶色表型RT-qPCR内参基因筛选及验证[J]. 林业科学研究, 2022, 35(1): 123-131. |
| Yang YY, Zhao LJ, Yang GJ, et al. Selection and validation of reference genes for leaf color phenotype in ‘maiyuanjinqiu’, a Catalpa fargesii variety, by RT-qPCR[J]. For Res, 2022, 35(1): 123-131. | |
| [22] |
He W, Xie R, Wang Y, et al. Comparative transcriptomic analysis on compatible/incompatible grafts in citrus[J]. Hortic Res, 2022, 9: uhab072.
doi: 10.1093/hr/uhab072 URL |
| [23] | 钟亚琴. 嫁接西瓜响应低钾胁迫的生理和分子机制研究[D]. 武汉: 华中农业大学, 2019. |
| Zhong YQ. Physiological and molecular mechanism of grafted watermelon in response to low potassium stress[D]. Wuhan: Huazhong Agricultural University, 2019. | |
| [24] | Thomas A, Brauer D, Sauer T, et al. Cultivar influences early rootstock and scion survival of grafted black walnut[J]. J Am Pomol Soc, 2008, 62(1): 3-12. |
| [25] |
Reig G, Zarrouk O, Font i Forcada C, et al. Anatomical graft compatibility study between apricot cultivars and different plum based rootstocks[J]. Sci Hortic, 2018, 237: 67-73.
doi: 10.1016/j.scienta.2018.03.035 URL |
| [26] | Huang Y, Zhao LQ, Kong QS, et al. Comprehensive mineral nutrition analysis of watermelon grafted onto two different rootstocks[J]. Hortic Plant J, 2016, 2(2): 105-113. |
| [27] | Bhuiyan MFA, Rahim MA, Alam MS. Study on the growth of plants produced by epicotyl(stone)grafting with different rootstock scion combinations in mango[J]. J Agrofor Environ, 2010, 3(2): 163-166. |
| [28] |
Harris ZN, Awale M, Bhakta N, et al. Multi-dimensional leaf phenotypes reflect root system genotype in grafted grapevine over the growing season[J]. GigaScience, 2021, 10(12): giab087.
doi: 10.1093/gigascience/giab087 URL |
| [29] | Yang YJ, Lu XM, Yan B, et al. Bottle gourd rootstock-grafting affects nitrogen metabolism in NaCl-stressed watermelon leaves and enhances short-term salt tolerance[J]. J Plant Physiol, 2013, 170(7): 653-661. |
| [30] |
Somkuwar RG, Bahetwar A, Khan I, et al. Changes in growth, photosynthetic activities, biochemical parameters and amino acid profile of Thompson Seedless grapes(Vitis vinifera L.)[J]. J Environ Biol, 2014, 35(6): 1157-1163.
pmid: 25522520 |
| [31] |
Proebsting WM, Maggard SP, Guo WW. The relationship of thiamine to the Alt locus of Pisum sativum L[J]. J Plant Physiol, 1990, 136(2): 231-235.
doi: 10.1016/S0176-1617(11)81671-8 URL |
| [32] |
Sayed SA, Gadallah MAA. Effects of shoot and root application of thiamin on salt-stressed sunflower plants[J]. Plant Growth Regul, 2002, 36(1): 71-80.
doi: 10.1023/A:1014784831387 URL |
| [33] |
Xu Q, Guo SR, Li L, et al. Proteomics analysis of compatibility and incompatibility in grafted cucumber seedlings[J]. Plant Physiol Biochem, 2016, 105: 21-28.
doi: 10.1016/j.plaphy.2016.04.001 URL |
| [34] |
Kim SH, Kim YH, Ahn YO, et al. Downregulation of the lycopene ϵ-cyclase gene increases carotenoid synthesis via the β-branch-specific pathway and enhances salt-stress tolerance in sweetpotato transgenic calli[J]. Physiol Plant, 2013, 147(4): 432-442.
doi: 10.1111/ppl.2013.147.issue-4 URL |
| [35] |
Komatsu A, Takanokura Y, Moriguchi T, et al. Differential expression of three sucrose-phosphate synthase isoforms during sucrose accumulation in citrus fruits(Citrus unshiu Marc.)[J]. Plant Science, 1999, 140(2): 169-178.
doi: 10.1016/S0168-9452(98)00217-9 URL |
| [36] |
Zhu YJ, Moore PH. Sucrose accumulation in the sugarcane stem is regulated by the difference between the activities of soluble acid invertase and sucrose phosphate synthase[J]. Plant Physiol, 1997, 115(2): 609-616.
doi: 10.1104/pp.115.2.609 pmid: 12223829 |
| [37] |
Pauly M, Keegstra K. Plant cell wall polymers as precursors for biofuels[J]. Curr Opin Plant Biol, 2010, 13(3): 304-311.
doi: 10.1016/j.pbi.2009.12.009 URL |
| [1] | LIN Hong-yan, GUO Xiao-rui, LIU Di, LI Hui, LU Hai. Molecular Mechanism of Transcriptional Factor AtbHLH68 in Regulating Cell Wall Development by Transcriptome Analysis [J]. Biotechnology Bulletin, 2023, 39(9): 105-116. |
| [2] | LOU Hui, ZHU Jin-cheng, YANG Yang, ZHANG Wei. Effects of Root Exudates in Resistant and Susceptible Varieties of Cotton on the Growths and Gene Expressions of Fusarium oxysporum [J]. Biotechnology Bulletin, 2023, 39(9): 156-167. |
| [3] | MIAO Yong-mei, MIAO Cui-ping, YU Qing-cai. Properties of Bacillus subtilis Strain BBs-27 Fermentation Broth and the Inhibition of Lipopeptides Against Fusarium culmorum [J]. Biotechnology Bulletin, 2023, 39(9): 255-267. |
| [4] | KONG De-zhen, DUAN Zhen-yu, WANG Gang, ZHANG Xin, XI Lin-qiao. Physiological Characteristics and Transcriptome Analysis of Sorghum bicolor × S. Sudanense Seedlings Under Salt-alkali Stress [J]. Biotechnology Bulletin, 2023, 39(6): 199-207. |
| [5] | YANG Yang, ZHU Jin-cheng, LOU Hui, HAN Ze-gang, ZHANG Wei. Transcriptome Analysis of Interaction Between Gossypium barbadense and Fusarium oxysporum f. sp. vasinfectum [J]. Biotechnology Bulletin, 2023, 39(6): 259-273. |
| [6] | LIU Hui, LU Yang, YE Xi-miao, ZHOU Shuai, LI Jun, TANG Jian-bo, CHEN En-fa. Comparative Transcriptome Analysis of Cadmium Stress Response Induced by Exogenous Sulfur in Tartary Buckwheat [J]. Biotechnology Bulletin, 2023, 39(5): 177-191. |
| [7] | XIE Yang, XING Yu-meng, ZHOU Guo-yan, LIU Mei-yan, YIN Shan-shan, YAN Li-ying. Transcriptome Analysis of Diploid and Autotetraploid in Cucumber Fruit [J]. Biotechnology Bulletin, 2023, 39(3): 152-162. |
| [8] | HU Li-li, LIN Bo-rong, WANG Hong-hong, CHEN Jian-song, LIAO Jin-ling, ZHUO Kan. Transcriptome and Candidate Effectors Analysis of Pratylenchus brachyurus [J]. Biotechnology Bulletin, 2023, 39(3): 254-266. |
| [9] | SUN Yan-qiu, XIE Cai-yun, TANG Yue-qin. Construction and Mechanism Analysis of High-temperature Resistant Saccharomyces cerevisiae [J]. Biotechnology Bulletin, 2023, 39(11): 226-237. |
| [10] | XU Jun, YE Yu-qing, NIU Ya-jing, HUANG He, ZHANG Meng-meng. Transcriptome Analysis of Rhizome Development in Chrysanthemum× × morifolium [J]. Biotechnology Bulletin, 2023, 39(10): 231-245. |
| [11] | LUO Hao-tian, WANG Long, WANG Yu-qian, WANG Yue, LI Jia-zhen, YANG Meng-ke, ZHANG Jie, DENG Xin, WANG Hong-yan. Genome-wide Identification and Expression Analysis of the RNAi-related Gene Families in Setaria viridis [J]. Biotechnology Bulletin, 2023, 39(1): 175-186. |
| [12] | XIN Jian-pan, LI Yan, ZHAO Chu, TIAN Ru-nan. Transcriptome Sequencing in the Leaves of Pontederia cordata with Cadmium Exposure and Gene Mining in Phenypropanoid Pathways [J]. Biotechnology Bulletin, 2022, 38(6): 198-210. |
| [13] | XU Jin, LI Tao, LI Chu-lin, ZHU Shun-ni, WANG Zhong-ming, XIANG Wen-zhou. Effects of Temperature on the Growth,Total Lipid and Eicosapentaenoic Acid Synthesis of Eustigmatos sp. [J]. Biotechnology Bulletin, 2022, 38(6): 261-271. |
| [14] | XIONG He-li, SHA Qian, LIU Shao-na, XIANG De-cai, ZHANG Bin, ZHAO Zhi-yong. Application of Single-cell Transcriptome Sequencing in Animals [J]. Biotechnology Bulletin, 2022, 38(3): 226-233. |
| [15] | ZHANG Bin, YANG Xin-xia. Identification of Key Transcription Factors in Response to Salt Stress in Rice [J]. Biotechnology Bulletin, 2022, 38(3): 9-15. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||