








Biotechnology Bulletin ›› 2023, Vol. 39 ›› Issue (9): 236-245.doi: 10.13560/j.cnki.biotech.bull.1985.2023-0022
Previous Articles Next Articles
HAN Hao-zhang( ), ZHANG Li-hua, LI Su-hua, ZHAO Rong, WANG Fang, WANG Xiao-li
), ZHANG Li-hua, LI Su-hua, ZHAO Rong, WANG Fang, WANG Xiao-li
Received:2023-01-11
Online:2023-09-26
Published:2023-10-24
HAN Hao-zhang, ZHANG Li-hua, LI Su-hua, ZHAO Rong, WANG Fang, WANG Xiao-li. Construction of cDNA Library of Cinnamomun bodinieri Induced by Saline-alkali Stress and Screening of CbP5CS Upstream Regulators[J]. Biotechnology Bulletin, 2023, 39(9): 236-245.
| 引物名称Primer name | 序列Sequence(5'-3') | 应用Application |
|---|---|---|
| CDS III/3' PCR primer | AAGCAGTGGTATCAACGCAGAGTGGCCATTATGGCCGGG | 逆转录 |
| SMART IV oligonucleotide | ATTCTAGAGGCCGAGGCGGCCGACATGTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTVN | 逆转录 |
| P1-F | CATATGGCCATGGAGGCCAGTGAATTCAAGCAGTGGTATCAACGCAGAGTGG | ds cDNA扩增 |
| P2-F | CATATGGCCATGGAGGCCAGTGAATTCAAAGCAGTGGTATCAACGCAGAGTGG | ds cDNA扩增 |
| P3-F | CATATGGCCATGGAGGCCAGTGAATTCAAAAGCAGTGGTATCAACGCAGAGTGG | ds cDNA扩增 |
| P4-R | CATCTGCAGCTCGAGCTCGATGGATCCCTAGAGGCCGAGGCGGCCGACATG | ds cDNA扩增 |
| Prime M1 | AAGCAGTGGTATCAACGCAGAGT | 均一化 |
| T7-F | taatacgactcactatagggcgag | 菌落PCR |
| AD-R | GCACGATGCACAGTTGAAG | 菌落PCR |
Table 1 Oligonucleotide primers used in this study
| 引物名称Primer name | 序列Sequence(5'-3') | 应用Application |
|---|---|---|
| CDS III/3' PCR primer | AAGCAGTGGTATCAACGCAGAGTGGCCATTATGGCCGGG | 逆转录 |
| SMART IV oligonucleotide | ATTCTAGAGGCCGAGGCGGCCGACATGTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTVN | 逆转录 |
| P1-F | CATATGGCCATGGAGGCCAGTGAATTCAAGCAGTGGTATCAACGCAGAGTGG | ds cDNA扩增 |
| P2-F | CATATGGCCATGGAGGCCAGTGAATTCAAAGCAGTGGTATCAACGCAGAGTGG | ds cDNA扩增 |
| P3-F | CATATGGCCATGGAGGCCAGTGAATTCAAAAGCAGTGGTATCAACGCAGAGTGG | ds cDNA扩增 |
| P4-R | CATCTGCAGCTCGAGCTCGATGGATCCCTAGAGGCCGAGGCGGCCGACATG | ds cDNA扩增 |
| Prime M1 | AAGCAGTGGTATCAACGCAGAGT | 均一化 |
| T7-F | taatacgactcactatagggcgag | 菌落PCR |
| AD-R | GCACGATGCACAGTTGAAG | 菌落PCR |

Fig. 2 Agarose gel electrophoresis of ds cDNA M: Marker. 1: ds cDNA amplified with P1-F/P4-R. 2: ds cDNA amplified with P2-F/P4-R. 3: ds cDNA amplified with P3-F/P4-R
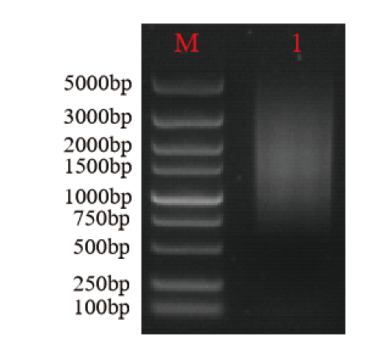
Fig. 3 Detection of normalization and removal of small fragments using agarose gel electrophoresis M: Marker. 1: Mixture of ds cDNA amplified with P1-F/P4-R, P2-F/P4-R, and P3-F/P4-R
| 位点 Site | 位置 Position | 序列 Sequence | 重复次数 Number of repetitions | 功能 Function |
|---|---|---|---|---|
| ABRE | -1 713 | ACGTG | 1 | 脱落酸响应元件 |
| AE-box | +191 | AGAAACAA | 1 | 部分光响应模块 |
| ARE | +680、-1 262、-693 | AAACCA | 3 | 厌氧诱导必需元件 |
| AT~TATA-box | +551 | TATATA | 1 | 光响应元件 |
| Box 4 | -1 846 | ATTAAT | 1 | 光响应中的保守DNA模块 |
| CAAT-box | +58、+75、-99、+139、+184、+196、+414、+513、+704、-743、-753、+770、+817、-819、+869、-902、+914、+941、+942、+1 005、-1 008、+1 115、 -1 118、1 140、-1 168、+1 195、+1 198、-1 275、 -1 293、+1 348、-1 351、-1 394、-1 429、+1 479、 +1 480、-1 498、+1 523、+1 540、+1 541、+1 570、+1 591、+1 617、-1 636、-1 813 | CAAAT/CAAT/CCAAT/TGCCAAC | 44 | 启动子和增强子区域普通顺式作用元件 |
| CAT-box | +1 976 | GCCACT | 1 | 分生组织表达响应元件 |
| CGTCA-motif | -143、-732、+146 | CGTCA | 3 | 茉莉酸甲酯响应的元件 |
| ERE | -1 743 | ATTTCATA | 1 | 乙烯响应元件 |
| G-Box | +1 713 | CACGTT | 1 | 光响应元件 |
| G-box | +80 | CACGAC | 1 | 光响应元件 |
| GT1-motif | -610、+1 225、+665 | GGTTAA/GTGTGTGAA | 3 | 光响应元件 |
| MYB | +2、-1 258、+1 201、-1 927、+1 001、-1 224 | CAACAG/CAACCA/TAACCA/ | 6 | MYB转录因子结合位点 |
| MYB-like sequence | -1 224、-1 927 | TAACCA | 2 | MYB转录因子结合位点 |
| MYC | -139、+817、-184 | CATTTG/CAATTG | 3 | bHLH转录因子结合位点 |
| Myb | -1 154、+1 485 | TAACTG | 2 | MYB转录因子结合位点 |
| Myb-binding site | +2、+1 201 | CAACAG | 2 | MYB转录因子结合位点 |
| P-box | -158 | CCTTTTG | 1 | 赤霉素响应元件 |
| SARE | -992 | TTCGACCATCTT | 1 | 水杨酸响应元件 |
| STRE | -265、+1 080、+1 801、+572、+1 962、-1 402、 -1 834、-281、-712 | AGGGG | 9 | 压力响应元件 |
| TATA-box | +202、-550、+267、+551、+552、+553、-1 161、 -1 162、+1 178、-1 179、-1 180、+1 471、-1 472、 -1 473、+1 554、-1 741、+1 850、-1 851、-1 917 | TATA/TATATAA/ccTATAAAaa/TATATA/ATATAA/ATATAT/ATTATA/TATAA/TACAAAA | 19 | -30位置附近核心启动子转录起始位点 |
| TATC-box | -258 | TATCCCA | 1 | 赤霉素响应元件 |
| TCA-element | -989、+1 408 | CCATCTTTTT | 2 | 水杨酸响应元件 |
| TCCC-motif | +1 870 | TCTCCCT | 1 | 光响应元件 |
| TGACG-motif | +143、+732、-146 | TGACG | 3 | 茉莉酸甲酯响应元件 |
| Unnamed | -234、+1 640、+256、-1 712 | CGTGG | 4 | 未知 |
| Unnamed | -628 | TCCACGTAGA | 1 | 未知 |
| Unnamed | -9、-1 083、+643、+1 871、-568、1 377、-833、 -1 191、-536、+1 863、+710、-635、-921 | CTCC | 13 | 未知 |
| WRE3 | -829、+1 865、-1 098 | CCACCT | 3 | 愈伤调节响应元件 |
| WUN-motif | -559 | AAATTACT | 1 | 愈伤调节响应元件 |
Table 2 cis-acting regulatory elements of the CbP5CS promoter in C. bodinieri
| 位点 Site | 位置 Position | 序列 Sequence | 重复次数 Number of repetitions | 功能 Function |
|---|---|---|---|---|
| ABRE | -1 713 | ACGTG | 1 | 脱落酸响应元件 |
| AE-box | +191 | AGAAACAA | 1 | 部分光响应模块 |
| ARE | +680、-1 262、-693 | AAACCA | 3 | 厌氧诱导必需元件 |
| AT~TATA-box | +551 | TATATA | 1 | 光响应元件 |
| Box 4 | -1 846 | ATTAAT | 1 | 光响应中的保守DNA模块 |
| CAAT-box | +58、+75、-99、+139、+184、+196、+414、+513、+704、-743、-753、+770、+817、-819、+869、-902、+914、+941、+942、+1 005、-1 008、+1 115、 -1 118、1 140、-1 168、+1 195、+1 198、-1 275、 -1 293、+1 348、-1 351、-1 394、-1 429、+1 479、 +1 480、-1 498、+1 523、+1 540、+1 541、+1 570、+1 591、+1 617、-1 636、-1 813 | CAAAT/CAAT/CCAAT/TGCCAAC | 44 | 启动子和增强子区域普通顺式作用元件 |
| CAT-box | +1 976 | GCCACT | 1 | 分生组织表达响应元件 |
| CGTCA-motif | -143、-732、+146 | CGTCA | 3 | 茉莉酸甲酯响应的元件 |
| ERE | -1 743 | ATTTCATA | 1 | 乙烯响应元件 |
| G-Box | +1 713 | CACGTT | 1 | 光响应元件 |
| G-box | +80 | CACGAC | 1 | 光响应元件 |
| GT1-motif | -610、+1 225、+665 | GGTTAA/GTGTGTGAA | 3 | 光响应元件 |
| MYB | +2、-1 258、+1 201、-1 927、+1 001、-1 224 | CAACAG/CAACCA/TAACCA/ | 6 | MYB转录因子结合位点 |
| MYB-like sequence | -1 224、-1 927 | TAACCA | 2 | MYB转录因子结合位点 |
| MYC | -139、+817、-184 | CATTTG/CAATTG | 3 | bHLH转录因子结合位点 |
| Myb | -1 154、+1 485 | TAACTG | 2 | MYB转录因子结合位点 |
| Myb-binding site | +2、+1 201 | CAACAG | 2 | MYB转录因子结合位点 |
| P-box | -158 | CCTTTTG | 1 | 赤霉素响应元件 |
| SARE | -992 | TTCGACCATCTT | 1 | 水杨酸响应元件 |
| STRE | -265、+1 080、+1 801、+572、+1 962、-1 402、 -1 834、-281、-712 | AGGGG | 9 | 压力响应元件 |
| TATA-box | +202、-550、+267、+551、+552、+553、-1 161、 -1 162、+1 178、-1 179、-1 180、+1 471、-1 472、 -1 473、+1 554、-1 741、+1 850、-1 851、-1 917 | TATA/TATATAA/ccTATAAAaa/TATATA/ATATAA/ATATAT/ATTATA/TATAA/TACAAAA | 19 | -30位置附近核心启动子转录起始位点 |
| TATC-box | -258 | TATCCCA | 1 | 赤霉素响应元件 |
| TCA-element | -989、+1 408 | CCATCTTTTT | 2 | 水杨酸响应元件 |
| TCCC-motif | +1 870 | TCTCCCT | 1 | 光响应元件 |
| TGACG-motif | +143、+732、-146 | TGACG | 3 | 茉莉酸甲酯响应元件 |
| Unnamed | -234、+1 640、+256、-1 712 | CGTGG | 4 | 未知 |
| Unnamed | -628 | TCCACGTAGA | 1 | 未知 |
| Unnamed | -9、-1 083、+643、+1 871、-568、1 377、-833、 -1 191、-536、+1 863、+710、-635、-921 | CTCC | 13 | 未知 |
| WRE3 | -829、+1 865、-1 098 | CCACCT | 3 | 愈伤调节响应元件 |
| WUN-motif | -559 | AAATTACT | 1 | 愈伤调节响应元件 |
| 基因ID Gene ID | 读数 Read count | 拟南芥同源序列 Arabidopsis thaliana homolog | E值E-value | 功能注释 Functional annotation |
|---|---|---|---|---|
| GW020491 | 1 | AT3G47120.1 | 8.00E-76 | 锌指 CCCH结构域蛋白25异构体 x1 |
| GW028183 | 15 | AT4G14410.2 | 6.00E-51 | AtbHLH104 |
| GW000650 | 3 | AT3G15420.1 | 1.00E-24 | 转录因子 TFIIIC |
| GW009089 | 7 | AT4G00755.2 | 6.00E-59 | F-box 家族蛋白 |
| GW009287 | 8 | AT1G64230.2 | 3.00E-83 | 泛素连接酶 |
| GW012884 | 24 | AT5G15730.2 | 2.00E-137 | 蛋白激酶超家族 |
| GW000705 | 24 | AT4G02340.1 | 2.00E-47 | α/β 水解酶超家族蛋白 |
| GW003120 | 4 | AT5G04480.1 | 0 | 葡萄糖基转移酶超家族蛋白 |
| GW004800 | 7 | AT5G63030.1 | 4.00E-42 | 硫氧还蛋白超家族蛋白 |
| GW006791 | 16 | ATMG01360.1 | 0 | 细胞色素氧化酶 |
| GW007525 | 13 | AT3G47550.6 | 6.00E-41 | RING/FYVE/PHD锌指超家族蛋白 |
| GW008185 | 15 | AT2G20360.1 | 1.00E-161 | NAD(P)结合罗斯曼卷曲超家族蛋白 |
| GW008711 | 19 | AT4G14305.1 | 8.00E-79 | 过氧化物酶体膜22 kD(Mpv17/PMP22)家族蛋白 |
| GW010514 | 3 | AT2G27030.3 | 2.00E-82 | 钙调素 |
| GW011043 | 24 | AT4G35450.1 | 3.00E-115 | 锚蛋白重复序列含有蛋白 |
| GW011868 | 16 | AT4G26220.1 | 3.00E-80 | S-腺苷甲硫氨酸依赖的甲基转移酶超家族蛋白 |
| GW011935 | 13 | AT1G55160.1 | 2.00E-35 | 功能未知的蛋白质 |
| GW012148 | 1 | AT1G54290.1 | 3.00E-48 | 翻译起始因子eIF1 |
| GW012186 | 19 | AT5G42850.2 | 4.00E-40 | 硫氧还蛋白超家族蛋白 |
| GW014171 | 56 | AT2G44860.2 | 2.00E-62 | 核糖体蛋白质 L24e 家族蛋白 |
| GW014289 | 34 | AT2G27020.1 | 1.00E-129 | 20s 蛋白酶体α亚基G1 |
| GW014358 | 24 | AT1G68590.1 | 1.00E-43 | 核糖体蛋白质 PSRP-3/Ycf65 |
| GW014549 | 10 | AT4G32140.1 | 4.00E-98 | 类EamA转运家族蛋白 |
| GW015686 | 1 | AT1G10200.1 | 2.00E-80 | GATA型锌指转录因子家族蛋白 |
| GW015796 | 64 | AT1G13950.1 | 4.00E-80 | 真核延伸因子5A-1 |
| GW017908 | 15 | AT3G55440.1 | 4.00E-111 | 磷酸丙糖异构酶 |
| GW020994 | 23 | AT5G53560.1 | 2.00E-48 | 细胞色素 B5亚型E |
| GW022429 | 11 | ATMG01360.1 | 0 | 细胞色素氧化酶 |
| GW024339 | 14 | AT4G19420.1 | 2.00E-153 | 果胶乙酰酯酶家族蛋白 |
| GW025798 | 12 | AT1G53680.1 | 1.00E-69 | 谷胱甘肽S-转移酶TAU28 |
| GW027120 | 11 | AT1G72210.1 | 4.00E-52 | AtbHLH96 |
Table 3 EST blast and annotation results
| 基因ID Gene ID | 读数 Read count | 拟南芥同源序列 Arabidopsis thaliana homolog | E值E-value | 功能注释 Functional annotation |
|---|---|---|---|---|
| GW020491 | 1 | AT3G47120.1 | 8.00E-76 | 锌指 CCCH结构域蛋白25异构体 x1 |
| GW028183 | 15 | AT4G14410.2 | 6.00E-51 | AtbHLH104 |
| GW000650 | 3 | AT3G15420.1 | 1.00E-24 | 转录因子 TFIIIC |
| GW009089 | 7 | AT4G00755.2 | 6.00E-59 | F-box 家族蛋白 |
| GW009287 | 8 | AT1G64230.2 | 3.00E-83 | 泛素连接酶 |
| GW012884 | 24 | AT5G15730.2 | 2.00E-137 | 蛋白激酶超家族 |
| GW000705 | 24 | AT4G02340.1 | 2.00E-47 | α/β 水解酶超家族蛋白 |
| GW003120 | 4 | AT5G04480.1 | 0 | 葡萄糖基转移酶超家族蛋白 |
| GW004800 | 7 | AT5G63030.1 | 4.00E-42 | 硫氧还蛋白超家族蛋白 |
| GW006791 | 16 | ATMG01360.1 | 0 | 细胞色素氧化酶 |
| GW007525 | 13 | AT3G47550.6 | 6.00E-41 | RING/FYVE/PHD锌指超家族蛋白 |
| GW008185 | 15 | AT2G20360.1 | 1.00E-161 | NAD(P)结合罗斯曼卷曲超家族蛋白 |
| GW008711 | 19 | AT4G14305.1 | 8.00E-79 | 过氧化物酶体膜22 kD(Mpv17/PMP22)家族蛋白 |
| GW010514 | 3 | AT2G27030.3 | 2.00E-82 | 钙调素 |
| GW011043 | 24 | AT4G35450.1 | 3.00E-115 | 锚蛋白重复序列含有蛋白 |
| GW011868 | 16 | AT4G26220.1 | 3.00E-80 | S-腺苷甲硫氨酸依赖的甲基转移酶超家族蛋白 |
| GW011935 | 13 | AT1G55160.1 | 2.00E-35 | 功能未知的蛋白质 |
| GW012148 | 1 | AT1G54290.1 | 3.00E-48 | 翻译起始因子eIF1 |
| GW012186 | 19 | AT5G42850.2 | 4.00E-40 | 硫氧还蛋白超家族蛋白 |
| GW014171 | 56 | AT2G44860.2 | 2.00E-62 | 核糖体蛋白质 L24e 家族蛋白 |
| GW014289 | 34 | AT2G27020.1 | 1.00E-129 | 20s 蛋白酶体α亚基G1 |
| GW014358 | 24 | AT1G68590.1 | 1.00E-43 | 核糖体蛋白质 PSRP-3/Ycf65 |
| GW014549 | 10 | AT4G32140.1 | 4.00E-98 | 类EamA转运家族蛋白 |
| GW015686 | 1 | AT1G10200.1 | 2.00E-80 | GATA型锌指转录因子家族蛋白 |
| GW015796 | 64 | AT1G13950.1 | 4.00E-80 | 真核延伸因子5A-1 |
| GW017908 | 15 | AT3G55440.1 | 4.00E-111 | 磷酸丙糖异构酶 |
| GW020994 | 23 | AT5G53560.1 | 2.00E-48 | 细胞色素 B5亚型E |
| GW022429 | 11 | ATMG01360.1 | 0 | 细胞色素氧化酶 |
| GW024339 | 14 | AT4G19420.1 | 2.00E-153 | 果胶乙酰酯酶家族蛋白 |
| GW025798 | 12 | AT1G53680.1 | 1.00E-69 | 谷胱甘肽S-转移酶TAU28 |
| GW027120 | 11 | AT1G72210.1 | 4.00E-52 | AtbHLH96 |
| [1] |
Xiao ZF, Jin ZN, Zhang BH, et al. Effects of IBA on rooting ability of Cinnamomum bodinieri citral type micro-shoots from transcriptomics analysis[J]. Plant Biotechnol Rep, 2020, 14(4): 467-477.
doi: 10.1007/s11816-020-00626-5 |
| [2] |
Guo HJ, Hu ZQ, Zhang HM, et al. Comparative effects of salt and alkali stress on antioxidant system in cotton(Gossypium hirsutum L.) leaves[J]. Open Chem, 2019, 17(1): 1352-1360.
doi: 10.1515/chem-2019-0147 URL |
| [3] |
Liu J, Wang YQ, Li QT. Analysis of differentially expressed genes and adaptive mechanisms of Prunus triloba Lindl. under alkaline stress[J]. Hereditas, 2017, 154: 10.
doi: 10.1186/s41065-017-0031-7 URL |
| [4] | Liu BS, Kang CL, Wang X, et al. Tolerance mechanisms of Leymus chinensis to salt-alkaline stress[J]. Acta Agric Scand Sect B Soil Plant Sci, 2015, 65(8): 723-734. |
| [5] |
Wang Y, Wang JC, Guo DD, et al. Physiological and comparative transcriptome analysis of leaf response and physiological adaption to saline alkali stress across pH values in alfalfa(Medicago sativa)[J]. Plant Physiol Biochem, 2021, 167: 140-152.
doi: 10.1016/j.plaphy.2021.07.040 URL |
| [6] |
Noreen S, Akhter MS, Yaamin T, et al. The ameliorative effects of exogenously applied proline on physiological and biochemical parameters of wheat(Triticum aestivum L.) crop under copper stress condition[J]. J Plant Interact, 2018, 13(1): 221-230.
doi: 10.1080/17429145.2018.1437480 URL |
| [7] |
Iqbal N. Photosynthetic differences in mustard genotypes under salinity stress: significance of proline metabolism[J]. Annu Res Rev Biol, 2014, 4: 3274-3296.
doi: 10.9734/ARRB URL |
| [8] |
Funck D, Baumgarten L, Stift M, et al. Differential contribution of P5CS isoforms to stress tolerance in Arabidopsis[J]. Front Plant Sci, 2020, 11: 565134.
doi: 10.3389/fpls.2020.565134 URL |
| [9] |
He L, Shi XX, Wang YM, et al. Arabidopsis ANAC069 binds to C[A/G]CG[T/G]sequences to negatively regulate salt and osmotic stress tolerance[J]. Plant Mol Biol, 2017, 93(4): 369-387.
doi: 10.1007/s11103-016-0567-3 URL |
| [10] |
Verma D, Jalmi SK, Bhagat PK, et al. A bHLH transcription factor, MYC2, imparts salt intolerance by regulating proline biosynthesis in Arabidopsis[J]. FEBS J, 2020, 287(12): 2560-2576.
doi: 10.1111/febs.v287.12 URL |
| [11] |
Jin C, Li KQ, Xu XY, et al. A novel NAC transcription factor, PbeNAC1, of Pyrus betulifolia confers cold and drought tolerance via interacting with PbeDREBs and activating the expression of stress-responsive genes[J]. Front Plant Sci, 2017, 8: 1049.
doi: 10.3389/fpls.2017.01049 URL |
| [12] |
Dai WS, Wang M, Gong XQ, et al. The transcription factor FcWRKY40 of Fortunella crassifolia functions positively in salt tolerance through modulation of ion homeostasis and proline biosynthesis by directly regulating SOS2 and P5CS1 homologs[J]. New Phytol, 2018, 219(3): 972-989.
doi: 10.1111/nph.2018.219.issue-3 URL |
| [13] | Liang JH, Zheng J, Wu Z, et al. Strawberry FaNAC2 enhances tolerance to abiotic stress by regulating proline metabolism[J]. Plants(Basel), 2020, 9(11): 1417. |
| [14] |
Zhang WH, Yang GY, Mu D, et al. An ethylene-responsive factor BpERF11 negatively modulates salt and osmotic tolerance in Betula platyphylla[J]. Sci Rep, 2016, 6: 23085.
doi: 10.1038/srep23085 |
| [15] |
Qin LP, Wang LQ, Guo Y, et al. An ERF transcription factor from Tamarix hispida, ThCRF1, can adjust osmotic potential and reactive oxygen species scavenging capability to improve salt tolerance[J]. Plant Sci, 2017, 265: 154-166.
doi: 10.1016/j.plantsci.2017.10.006 URL |
| [16] |
Zhang H, Liu XL, Zhang RX, et al. Root damage under alkaline stress is associated with reactive oxygen species accumulation in rice(Oryza sativa L.)[J]. Front Plant Sci, 2017, 8: 1580.
doi: 10.3389/fpls.2017.01580 pmid: 28943882 |
| [17] | Legocka J, Sobieszczuk-Nowicka E, Ludwicki D, et al. Putrescine catabolism via DAO contributes to proline and GABA accumulation in roots of lupine seedlings growing under salt stress[J]. Acta Soc Bot Pol, 2017, 86(3): 3549. |
| [18] |
Liu LJ, Huang L, Lin XY, et al. Hydrogen peroxide alleviates salinity-induced damage through enhancing proline accumulation in wheat seedlings[J]. Plant Cell Rep, 2020, 39(5): 567-575.
doi: 10.1007/s00299-020-02513-3 pmid: 32025801 |
| [19] |
Ghosh UK, Islam MN, Siddiqui MN, et al. Proline, a multifaceted signaling molecule in plant responses to abiotic stress: understanding the physiological mechanisms[J]. Plant Biol J, 2022, 24(2): 227-239.
doi: 10.1111/plb.v24.2 URL |
| [20] |
Han GL, Wang MJ, Yuan F, et al. The CCCH zinc finger protein gene AtZFP1 improves salt resistance in Arabidopsis thaliana[J]. Plant Mol Biol, 2014, 86(3): 237-253.
doi: 10.1007/s11103-014-0226-5 URL |
| [21] |
Huang J, Sun SJ, Xu DQ, et al. A TFIIIA-type zinc finger protein confers multiple abiotic stress tolerances in transgenic rice(Oryza sativa L.)[J]. Plant Mol Biol, 2012, 80(3): 337-350.
doi: 10.1007/s11103-012-9955-5 pmid: 22930448 |
| [22] |
Yang TR, Yao SF, Hao L, et al. Wheat bHLH-type transcription factor gene TabHLH1 is crucial in mediating osmotic stresses tolerance through modulating largely the ABA-associated pathway[J]. Plant Cell Rep, 2016, 35(11): 2309-2323.
doi: 10.1007/s00299-016-2036-5 URL |
| [23] |
Guo MX, Li SP, Tian S, et al. Transcriptome analysis of genes involved in defense against alkaline stress in roots of wild jujube(Ziziphus acidojujuba)[J]. PLoS One, 2017, 12(10): e0185732.
doi: 10.1371/journal.pone.0185732 URL |
| [24] |
Qian YC, Zhang TY, Yu Y, et al. Regulatory mechanisms of bHLH transcription factors in plant adaptive responses to various abiotic stresses[J]. Front Plant Sci, 2021, 12: 677611.
doi: 10.3389/fpls.2021.677611 URL |
| [25] |
Wang FB, Zhu H, Kong WL, et al. The Antirrhinum AmDEL gene enhances flavonoids accumulation and salt and drought tolerance in transgenic Arabidopsis[J]. Planta, 2016, 244(1): 59-73.
doi: 10.1007/s00425-016-2489-3 URL |
| [26] |
Zhang HF, Guo JB, Chen XQ, et al. Pepper bHLH transcription factor CabHLH035 contributes to salt tolerance by modulating ion homeostasis and proline biosynthesis[J]. Hortic Res, 2022, 9: uhac203.
doi: 10.1093/hr/uhac203 URL |
| [1] | HUANG Xiao-long, SUN Gui-lian, MA Dan-dan, YAN Hui-qing. Construction of Yeast One-hybrid Library and Screening of Factors Regulating LAZY1 Expression in Rice [J]. Biotechnology Bulletin, 2023, 39(9): 126-135. |
| [2] | LYU Qiu-yu, SUN Pei-yuan, RAN Bin, WANG Jia-rui, CHEN Qing-fu, LI Hong-you. Cloning, Subcellular Localization and Expression Analysis of the Transcription Factor Gene FtbHLH3 in Fagopyrum tataricum [J]. Biotechnology Bulletin, 2023, 39(8): 194-203. |
| [3] | XU Jing, ZHU Hong-lin, LIN Yan-hui, TANG Li-qiong, TANG Qing-jie, WANG Xiao-ning. Cloning of IbHQT1 Promoter and Identification of Upstream Regulatory Factors in Sweet Potato [J]. Biotechnology Bulletin, 2023, 39(8): 213-219. |
| [4] | LI Bo, LIU He-xia, CHEN Yu-ling, ZHOU Xing-wen, ZHU Yu-lin. Cloning, Subcellular Localization and Expression Analysis of CnbHLH79 Transcription Factor from Camellia nitidissima [J]. Biotechnology Bulletin, 2023, 39(8): 241-250. |
| [5] | CHEN Xiao, YU Ming-lan, WU Long-kun, ZHENG Xiao-ming, PANG Hong-bo. Research Progress in lncRNA and Their Responses to Low Temperature Stress in Plant [J]. Biotechnology Bulletin, 2023, 39(7): 1-12. |
| [6] | GUO Yi-ting, ZHAO Wen-ju, REN Yan-jing, ZHAO Meng-liang. Identification and Analysis of NAC Transcription Factor Family Genes in Helianthus tuberosus L. [J]. Biotechnology Bulletin, 2023, 39(6): 217-232. |
| [7] | FENG Shan-shan, WANG Lu, ZHOU Yi, WANG You-ping, FANG Yu-jie. Research Progresses on WOX Family Genes in Regulating Plant Development and Abiotic Stress Response [J]. Biotechnology Bulletin, 2023, 39(5): 1-13. |
| [8] | WANG Bing, ZHAO Hui-na, YU Jing, YU Shi-zhou, LEI Bo. Research Progress in the Regulation of Plant Branch Development [J]. Biotechnology Bulletin, 2023, 39(5): 14-22. |
| [9] | ZHANG Xin-bo, CUI Hao-liang, SHI Pei-hua, GAO Jin-chun, ZHAO Shun-ran, TAO Chen-yu. Research Progress in Low-input Chromatin Immunoprecipitation Assay [J]. Biotechnology Bulletin, 2023, 39(4): 227-235. |
| [10] | GE Yan-rui, ZHAO Ran, XU Jing, LI Ruo-fan, HU Yun-tao, LI Rui-li. Advances in the Development and Regulation of Vascular Cambium [J]. Biotechnology Bulletin, 2023, 39(3): 13-25. |
| [11] | LIU Cheng-xia, SUN Zong-yan, LUO Yun-bo, ZHU Hong-liang, QU Gui-qin. Multifaceted Roles of bHLH Phosphorylation in Regulation of Plant Physiological Functions [J]. Biotechnology Bulletin, 2023, 39(3): 26-34. |
| [12] | ZHAO Meng-liang, GUO Yi-ting, REN Yan-jing. Identification and Analysis of WRKY Transcription Factor Family Genes in Helianthus tuberosus [J]. Biotechnology Bulletin, 2023, 39(2): 116-125. |
| [13] | HAN Fang-ying, HU Xin, WANG Nan-nan, XIE Yu-hong, WANG Xiao-yan, ZHU Qiang. Research Progress in Response of DREBs to Abiotic Stress in Plant [J]. Biotechnology Bulletin, 2023, 39(11): 86-98. |
| [14] | CHEN Chu-yi, YANG Xiao-mei, CHEN Sheng-yan, CHEN Bin, YUE Li-ran. Expression Analysis of the ZF-HD Gene Family in Chrysanthemum nankingense Under Drought and ABA Treatment [J]. Biotechnology Bulletin, 2023, 39(11): 270-282. |
| [15] | FENG Ce-ting, JIANG Lyu, LIU Xing-ying, LUO Le, PAN Hui-tang, ZHANG Qi-xiang, YU Chao. Identification of the NAC Gene Family in Rosa persica and Response Analysis Under Drought Stress [J]. Biotechnology Bulletin, 2023, 39(11): 283-296. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||