








Biotechnology Bulletin ›› 2024, Vol. 40 ›› Issue (10): 253-261.doi: 10.13560/j.cnki.biotech.bull.1985.2024-0301
Previous Articles Next Articles
JIANG Yu-shan1( ), LAN Qian1, WANG Fang1, JIANG Liang1, PEI Cheng-cheng1,2(
), LAN Qian1, WANG Fang1, JIANG Liang1, PEI Cheng-cheng1,2( )
)
Received:2024-03-27
Online:2024-10-26
Published:2024-11-20
Contact:
PEI Cheng-cheng
E-mail:jiangyushan99@163.com;2304609281@sxau.edu.cn
JIANG Yu-shan, LAN Qian, WANG Fang, JIANG Liang, PEI Cheng-cheng. Characterization of a Quinoa Mutant Affecting Tyrosine Metabolism[J]. Biotechnology Bulletin, 2024, 40(10): 253-261.

Fig. 1 Phenotypic comparison between ‘Red Quinoa 1’(RQ1)and its mutant(gq1)during different development stages A: 2-week-old seedling; B: side view of 2-week-old seedling; C: plants during the filling stage; D: young panicle of grain filling stage; E: seed. Scale bar: 1 cm for A, B, D, and E, respectively, and 10 cm for C
| 杂交组合 Hybrid combination of F2 | 总计 Total | 红色单株个体数 Number of red individuals | 绿色单株个体数 Number of green individuals | x2(3∶1) | P |
|---|---|---|---|---|---|
| RQ1/gq1 | 163 | 128 | 35 | 1.082 | 0.298 |
Table 1 Test of Chi-square on segregation rate of F2 population between gq1 and RQ1
| 杂交组合 Hybrid combination of F2 | 总计 Total | 红色单株个体数 Number of red individuals | 绿色单株个体数 Number of green individuals | x2(3∶1) | P |
|---|---|---|---|---|---|
| RQ1/gq1 | 163 | 128 | 35 | 1.082 | 0.298 |
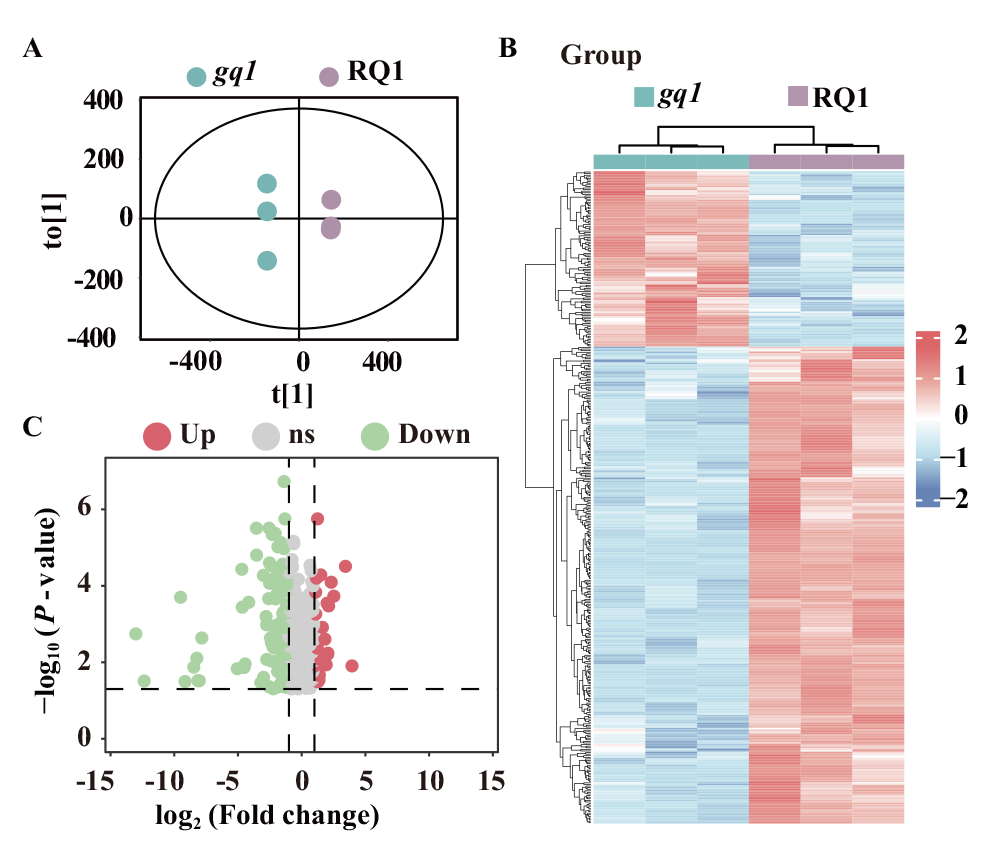
Fig. 2 Overview of differential metabolite analysis in gq1 vs RQ1 A: Score plots of the OPLS-DA model for gq1 vs RQ1. B: Heatmap of hierarchical clustering analysis for differential metabolites. C: Volcano plot of differential metabolites based on fold change
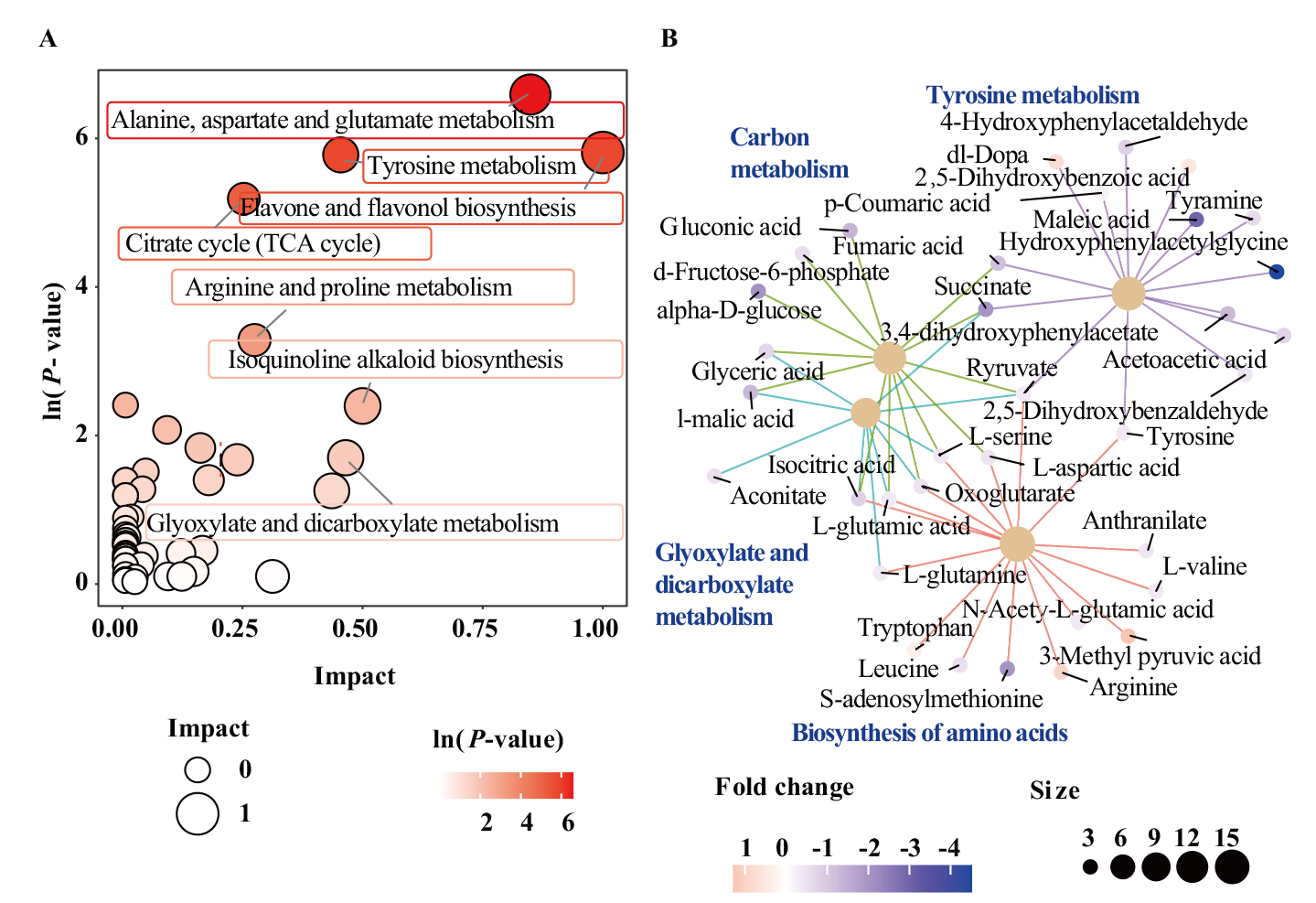
Fig. 4 KEGG pathway enrichment analysis of differential metabolites between RQ1 and gq1 A: KEGG pathway enrichment analysis of qg1 vs RQ1; B: KEGG category netplot of qg1 vs RQ1
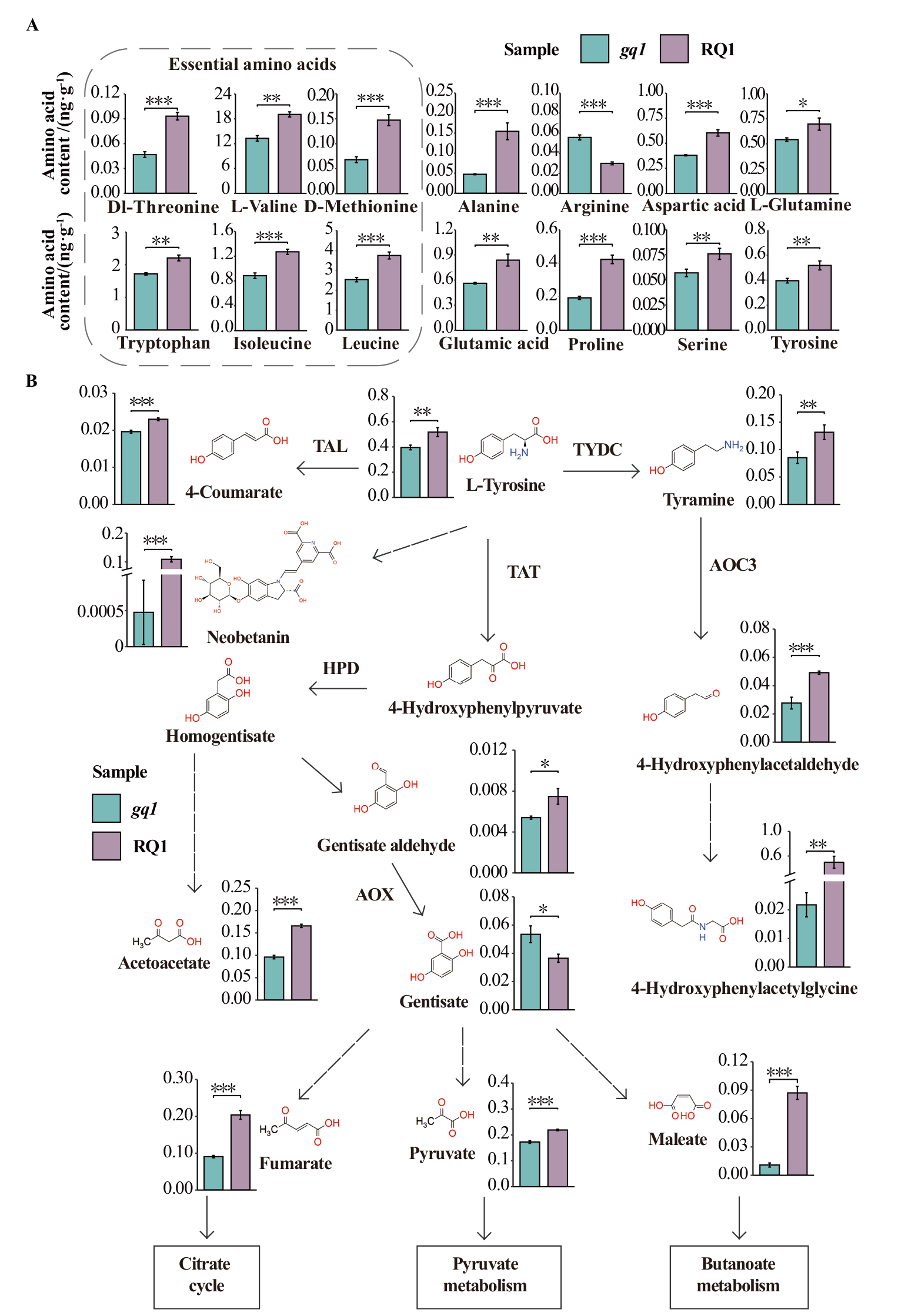
Fig. 5 Variation of amino acid content in mutant gq1 A: Contents of 14 differentially expressed amino acids in gq1 and RQ1. B: An overview of metabolites and their expressions in tyrosine metabolism pathway. Arrows with dashed lines designate multiple enzymes steps. TAT: Tyrosine aminotransferase; TAL: tyrosine ammonia-lyase; TYDC: L-tyrosine decarboxylase; HPD: hydroxyphenylpyruvate dioxygenase; AOC3: primary-amine oxidase; AOX: aldehyde oxidase. Variables of significance(* P ≤ 0.05, ** P ≤ 0.01, *** P ≤ 0.001)
| [1] | Alandia G, Rodriguez JP, Jacobsen SE, et al. Global expansion of quinoa and challenges for the Andean region[J]. Glob Food Secur, 2020, 26: 100429. |
| [2] | Abd El-Samad EH, Hussin SA, El-Naggar AM, et al. The potential use of quinoa as a new non-traditional leafy vegetable crop[J]. Bioscience Research, 2018, 15(4): 3387-3403. |
| [3] |
Filho AMM, Pirozi MR, Borges JT, et al. Quinoa: nutritional, functional, and antinutritional aspects[J]. Crit Rev Food Sci Nutr, 2017, 57(8): 1618-1630.
doi: 10.1080/10408398.2014.1001811 pmid: 26114306 |
| [4] |
Abugoch James LE. Quinoa(Chenopodium quinoa Willd.): composition, chemistry, nutritional, and functional properties[J]. Adv Food Nutr Res, 2009, 58: 1-31.
doi: 10.1016/S1043-4526(09)58001-1 pmid: 19878856 |
| [5] | Adolf VI, Shabala S, Andersen MN, et al. Varietal differences of quinoa's tolerance to saline conditions[J]. Plant Soil, 2012, 357(1): 117-129. |
| [6] | Sun WJ, Yao M, Wang Z, et al. Involvement of auxin-mediated CqEXPA50 contributes to salt tolerance in quinoa(Chenopodium quinoa)by interaction with auxin pathway genes[J]. Int J Mol Sci, 2022, 23(15): 8480. |
| [7] | Sun WJ, Wei JL, Wu GM, et al. CqZF-HD14 enhances drought tolerance in quinoa seedlings through interaction with CqHIPP34 and CqNAC79[J]. Plant Sci, 2022, 323: 111406. |
| [8] | Hinojosa L, González JA, Barrios-Masias FH, et al. Quinoa abiotic stress responses: a review[J]. Plants, 2018, 7(4): 106. |
| [9] | Jacobsen SE, Monteros C, Corcuera LJ, et al. Frost resistance mechanisms in quinoa(Chenopodium quinoa Willd.)[J]. Eur J Agron, 2007, 26(4): 471-475. |
| [10] | Ain QT, Siddique K, Bawazeer S, et al. Adaptive mechanisms in quinoa for coping in stressful environments: an update[J]. PeerJ, 2023, 11: e14832. |
| [11] | Graf BL, Rojas-Silva P, Rojo LE, et al. Innovations in health value and functional food development of quinoa(Chenopodium quinoa Willd.)[J]. Compr Rev Food Sci Food Saf, 2015, 14(4): 431-445. |
| [12] | Carciochi RA, Galván-D'Alessandro L, Vandendriessche P, et al. Effect of germination and fermentation process on the antioxidant compounds of quinoa seeds[J]. Plant Foods Hum Nutr, 2016, 71(4): 361-367. |
| [13] | Escuredo O, González Martín MI, Wells Moncada G, et al. Amino acid profile of the quinoa(Chenopodium quinoa Willd.) using near infrared spectroscopy and chemometric techniques[J]. J Cereal Sci, 2014, 60(1): 67-74. |
| [14] | Repo-Carrasco R, Espinoza C, Jacobsen SE. Nutritional value and use of the Andean crops quinoa(Chenopodium quinoa)and kañiwa(Chenopodium pallidicaule)[J]. Food Rev Int, 2003, 19(1/2): 179-189. |
| [15] | Guo HM, Hao YQ, Yang XS, et al. Exploration on bioactive properties of quinoa protein hydrolysate and peptides: a review[J]. Crit Rev Food Sci Nutr, 2023, 63(16): 2896-2909. |
| [16] |
Tang Y, Li XH, Chen PX, et al. Characterisation of fatty acid, carotenoid, tocopherol/tocotrienol compositions and antioxidant activities in seeds of three Chenopodium quinoa Willd. genotypes[J]. Food Chem, 2015, 174: 502-508.
doi: 10.1016/j.foodchem.2014.11.040 pmid: 25529712 |
| [17] | Chen X, Zhang YY, Cao BE, et al. Assessment and comparison of nutritional qualities of thirty quinoa(Chenopodium quinoa Willd.) seed varieties[J]. Food Chem X, 2023, 19: 100808. |
| [18] | Adamczewska-Sowińska K, Sowiński J, Jama-Rodzeńska A. The effect of sowing date and harvest time on leafy greens of quinoa(Chenopodium quinoa Willd.) yield and selected nutritional parameters[J]. Agriculture, 2021, 11(5): 405. |
| [19] | Pathan S, Eivazi F, Valliyodan B, et al. Nutritional composition of the green leaves of quinoa(Chenopodium quinoa Willd.)[J]. J Food Res, 2019, 8(6): 55. |
| [20] | Pathan S, Siddiqui RA. Nutritional composition and bioactive components in quinoa(Chenopodium quinoa Willd.) greens: a review[J]. Nutrients, 2022, 14(3): 558. |
| [21] | Qian GT, Li XY, Zhang H, et al. Metabolomics analysis reveals the accumulation patterns of flavonoids and phenolic acids in quinoa(Chenopodium quinoa Willd.) grains of different colors[J]. Food Chem X, 2023, 17: 100594. |
| [22] | Liu JN, Liu J, Zhang P, et al. Elucidating the differentiation synthesis mechanisms of differently colored resistance quinoa seedings using metabolite profiling and transcriptome analysis[J]. Metabolites, 2023, 13(10): 1065. |
| [23] | Liu YJ, Liu JN, Kong ZY, et al. Transcriptomics and metabolomics analyses of the mechanism of flavonoid synthesis in seeds of differently colored quinoa strains[J]. Genomics, 2022, 114(1): 138-148. |
| [24] | Liu YJ, Liu JN, Li L, et al. Transcriptome and metabolome combined to analyze quinoa grain quality differences of different colors cultivars[J]. Int J Mol Sci, 2022, 23(21): 12883. |
| [25] | Cai YP, Weng K, Guo Y, et al. An integrated targeted metabolomic platform for high-throughput metabolite profiling and automated data processing[J]. Metabolomics, 2015, 11(6): 1575-1586. |
| [26] |
Smith CA, Want EJ, O'Maille G, et al. XCMS: processing mass spectrometry data for metabolite profiling using nonlinear peak alignment, matching, and identification[J]. Anal Chem, 2006, 78(3): 779-787.
doi: 10.1021/ac051437y pmid: 16448051 |
| [27] | Trygg J, Wold S. Orthogonal projections to latent structures(O-PLS)[J]. J Chemom, 2002, 16(3): 119-128. |
| [28] |
Gu ZG, Eils R, Schlesner M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data[J]. Bioinformatics, 2016, 32(18): 2847-2849.
doi: 10.1093/bioinformatics/btw313 pmid: 27207943 |
| [29] | Wu TZ, Hu EQ, Xu SB, et al. clusterProfiler 4.0: a universal enrichment tool for interpreting omics data[J]. Innovation(Camb), 2021, 2(3): 100141. |
| [30] |
Galili G, Amir R, Fernie AR. The regulation of essential amino acid synthesis and accumulation in plants[J]. Annu Rev Plant Biol, 2016, 67: 153-178.
doi: 10.1146/annurev-arplant-043015-112213 pmid: 26735064 |
| [31] |
Maeda H, Dudareva N. The shikimate pathway and aromatic amino acid biosynthesis in plants[J]. Annu Rev Plant Biol, 2012, 63: 73-105.
doi: 10.1146/annurev-arplant-042811-105439 pmid: 22554242 |
| [32] |
Sandell FL, Holzweber T, Street NR, et al. Genomic basis of seed colour in quinoa inferred from variant patterns using extreme gradient boosting[J]. Plant Biotechnol J, 2024, 22(5): 1312-1324.
doi: 10.1111/pbi.14267 pmid: 38213076 |
| [1] | LIN Tong, YUAN Cheng, DONG Chen-wen-hua, ZENG Meng-qiong, YANG Yan, MAO Zi-chao, LIN Chun. Screening and Functional Analysis of Gene CqSTK Associated with Gametophyte Development of Quinoa [J]. Biotechnology Bulletin, 2024, 40(8): 83-94. |
| [2] | SUN Hui-qiong, ZHANG Chun-lai, WANG Xi-liang, XU Hong-shen, DOU Miao-miao, YANG Bo-hui, CHAI Wen-ting, ZHAO Shan-shan, JIANG Xiao-dong. Identification, Expression and DNA Variation Analysis of FLS Gene Family in Chenopodium quinoa [J]. Biotechnology Bulletin, 2024, 40(7): 172-182. |
| [3] | HAN Le-le, SONG Wen-di, BIAN Jia-shen, LI Yang, YANG Shuang-sheng, CHEN Zi-yi, LI Xiao-wei. Revealing the Flavonoid Biosynthesis of Soybean GmERD15c under Salt Stress by Combined Analysis of Transcriptome and Metabolome [J]. Biotechnology Bulletin, 2024, 40(10): 243-252. |
| [4] | JI Hong-chao, LI Zheng-yan. Research Progress and Prospects in the Structural Annotation of Unknown Secondary Metabolites Based on Mass Spectrometry [J]. Biotechnology Bulletin, 2024, 40(10): 76-85. |
| [5] | HE Shi-yu, ZENG Zhong-da, LI Bo-yan. Application Progress of Spatially Resolved Metabolomics in Disease Diagnosis Research [J]. Biotechnology Bulletin, 2024, 40(1): 145-159. |
| [6] | ZHOU Ai-ting, PENG Rui-qi, WANG Fang, WU Jian-rong, MA Huan-cheng. Analysis of Metabolic Differences of Biocontrol Strain DZY6715 at Different Growth Stages [J]. Biotechnology Bulletin, 2023, 39(9): 225-235. |
| [7] | HAN Hua-rui, YANG Yu-lu, MEN Yi-han, HAN Shang-ling, HAN Yuan-huai, HUO Yi-qiong, HOU Si-yu. SiYABBYs Involved in Rhamnoside Biosynthesis During the Flower Development of Setaria italica, Based on Metabolomics [J]. Biotechnology Bulletin, 2023, 39(6): 189-198. |
| [8] | XU Yang, DING Hong, ZHANG Guan-chu, GUO Qing, ZHANG Zhi-meng, DAI Liang-xiang. Metabolomics Analysis of Germinating Peanut Seed Under Salt Stress [J]. Biotechnology Bulletin, 2023, 39(1): 199-213. |
| [9] | WEI Xin-xin, LAN Hai-yan. Advances in the Regulation of Plant MYB Transcription Factors in Secondary Metabolism and Stress Response [J]. Biotechnology Bulletin, 2022, 38(8): 12-23. |
| [10] | GULJAMAL·Aisa , XING Jun, LI An, ZHANG Rui. Non-targeted Metabolomics Analysis of Benzo(α)pyrene by Microorganisms in Kefir Grains [J]. Biotechnology Bulletin, 2022, 38(5): 123-135. |
| [11] | YANG Yu-ping, ZHANG Xia, WANG Chong-chong, WANG Xiao-yan. Study on Urine Metabolomics in Rats of Different Ages [J]. Biotechnology Bulletin, 2022, 38(2): 166-172. |
| [12] | ZHANG Ye-meng, ZHU Li-li, CHEN Zhi-guo. Identification and Expression Analysis of NHX Gene Family in Quinoa Under Salt Stress [J]. Biotechnology Bulletin, 2022, 38(12): 184-193. |
| [13] | WU Yu-ping, ZHOU Yong, PU Juan, LI Hui, ZHANG Jin-gang, ZHU Yan-ping. Application Progress of Metabolomics in Tumor Drug Target Screening [J]. Biotechnology Bulletin, 2022, 38(1): 311-318. |
| [14] | ZHANG Feng, CHEN Wei. Research Progress of Metabolomics in Plant Stress Biology [J]. Biotechnology Bulletin, 2021, 37(8): 1-11. |
| [15] | CAI Guo-lei, LU Xiao-kai, LOU Shui-zhu, YANG Hai-ying, DU Gang. Classification and Identification of Bacillus LM Based on Whole Genome and Study on Its Antibacterial Principle [J]. Biotechnology Bulletin, 2021, 37(8): 176-185. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||