








Biotechnology Bulletin ›› 2024, Vol. 40 ›› Issue (10): 243-252.doi: 10.13560/j.cnki.biotech.bull.1985.2024-0373
Previous Articles Next Articles
HAN Le-le1,2( ), SONG Wen-di1, BIAN Jia-shen1, LI Yang1, YANG Shuang-sheng1, CHEN Zi-yi1, LI Xiao-wei1,2(
), SONG Wen-di1, BIAN Jia-shen1, LI Yang1, YANG Shuang-sheng1, CHEN Zi-yi1, LI Xiao-wei1,2( )
)
Received:2024-04-18
Online:2024-10-26
Published:2024-11-20
Contact:
LI Xiao-wei
E-mail:1491410856@qq.com;xiaoweili1206@163.com
HAN Le-le, SONG Wen-di, BIAN Jia-shen, LI Yang, YANG Shuang-sheng, CHEN Zi-yi, LI Xiao-wei. Revealing the Flavonoid Biosynthesis of Soybean GmERD15c under Salt Stress by Combined Analysis of Transcriptome and Metabolome[J]. Biotechnology Bulletin, 2024, 40(10): 243-252.
| 代谢物级别 Metabolite level | 组别 Group | 代谢物数量 Number of metabolites | 上调数量 Up-regulated number | 下调数量 Down-regulated number | 总差异代谢物 Total differential metabolites |
|---|---|---|---|---|---|
| 一级差异代谢物 Primary differential metabolites | WT-mock VS OE-mock | 20 804 | 846 | 847 | 1 693 |
| WT-NaCl VS OE-NaCl | 20 804 | 840 | 735 | 1 575 | |
| 二级差异代谢物 Secondary differential metabolites | WT-mock VS OE-mock | 402 | 20 | 15 | 35 |
| WT-NaCl VS OE-NaCl | 402 | 31 | 21 | 52 |
Table 1 Differential metabolites
| 代谢物级别 Metabolite level | 组别 Group | 代谢物数量 Number of metabolites | 上调数量 Up-regulated number | 下调数量 Down-regulated number | 总差异代谢物 Total differential metabolites |
|---|---|---|---|---|---|
| 一级差异代谢物 Primary differential metabolites | WT-mock VS OE-mock | 20 804 | 846 | 847 | 1 693 |
| WT-NaCl VS OE-NaCl | 20 804 | 840 | 735 | 1 575 | |
| 二级差异代谢物 Secondary differential metabolites | WT-mock VS OE-mock | 402 | 20 | 15 | 35 |
| WT-NaCl VS OE-NaCl | 402 | 31 | 21 | 52 |

Fig. 3 Metabolite variation in flavonoid synthesis pathway of soybean with transgenic GmERD15c gene A: Main classification of secondary differential metabolites. B: KEGG analysis of transgenic soybean metabolites. C: Quantitative analysis of transgenic soybean flavonoid compounds
| 对照组Control | 实验组Treat | 上调表达基因Up-regulated genes | 下调表达基因Down-regulated genes | 差异表达基因Total DEGs |
|---|---|---|---|---|
| WT-mock | OE-mock | 605 | 1 208 | 1 813 |
| WT-NaCl | OE-NaCl | 681 | 693 | 1 374 |
| OE-mock | OE-NaCl | 5 348 | 4 456 | 9 804 |
| WT-mock | WT-NaCl | 6 576 | 6 533 | 13 109 |
Table 2 Transcriptome differential gene analysis in soybean
| 对照组Control | 实验组Treat | 上调表达基因Up-regulated genes | 下调表达基因Down-regulated genes | 差异表达基因Total DEGs |
|---|---|---|---|---|
| WT-mock | OE-mock | 605 | 1 208 | 1 813 |
| WT-NaCl | OE-NaCl | 681 | 693 | 1 374 |
| OE-mock | OE-NaCl | 5 348 | 4 456 | 9 804 |
| WT-mock | WT-NaCl | 6 576 | 6 533 | 13 109 |

Fig. 4 Upset diagram of transcriptome differential genes in soybean Number in each set indicates the number of all differential genes identified in each comparison group. Number of each intersect indicates the number of common differences identified by multiple comparison groups, a point on the abscissa indicates the number of unique differences identified by the comparison group, and a line connecting multiple points on the abscissa indicates the number of common differences identified by multiple comparison groups connected by the line. Blue indicates one group, yellow indicates two groups, red indicates three groups, and green indicates four groups
| 通路ID Pathway ID | KEGG通路 KEGG pathway | 1级通路 Level 1 | 2级通路 Level 2 | 上调数量 Up_number | 下调数量 Down_number | 差异数量 DEG_number | 总数量 Total_number |
|---|---|---|---|---|---|---|---|
| gmx00910 | 氮代谢 | 新陈代谢 | 能量代谢 | 6 | 2 | 8 | 44 |
| gmx00260 | 甘氨酸、丝氨酸和苏氨酸代谢 | 新陈代谢 | 氨基酸代谢 | 4 | 4 | 8 | 89 |
| gmx00940 | 苯丙烷类生物合成 | 新陈代谢 | 其他次生代谢产物的生物合成 | 8 | 7 | 15 | 260 |
| gmx00565 | 乙醚脂质代谢 | 新陈代谢 | 脂类代谢 | 5 | 0 | 5 | 43 |
| gmx00904 | 二萜类生物合成 | 新陈代谢 | 萜类化合物和聚酮类化合物的代谢 | 1 | 4 | 5 | 45 |
| gmx00564 | 甘油磷脂代谢 | 新陈代谢 | 脂类代谢 | 9 | 1 | 10 | 164 |
| gmx00592 | 亚麻酸代谢 | 新陈代谢 | 脂类代谢 | 4 | 2 | 6 | 72 |
| gmx00908 | 玉米素生物合成 | 新陈代谢 | 萜类化合物和聚酮类化合物的代谢 | 3 | 1 | 4 | 36 |
| gmx00350 | 酪氨酸代谢 | 新陈代谢 | 氨基酸代谢 | 3 | 2 | 5 | 56 |
| gmx00270 | 半胱氨酸和蛋氨酸代谢 | 新陈代谢 | 氨基酸代谢 | 6 | 3 | 9 | 154 |
| gmx00630 | 乙醛酸盐和二羧酸代谢 | 新陈代谢 | 碳水化合物代谢 | 0 | 6 | 6 | 87 |
| gmx00130 | 泛醌和其他萜类醌生物合成 | 新陈代谢 | 辅因子和维生素的代谢 | 4 | 1 | 5 | 68 |
| gmx00943 | 异黄酮生物合成 | 新陈代谢 | 其他次生代谢产物的生物合成 | 3 | 0 | 3 | 28 |
| gmx00941 | 类黄酮生物合成 | 新陈代谢 | 其他次生代谢产物的生物合成 | 3 | 2 | 5 | 77 |
Table 3 KEGG analysis of soybean transcriptome transgenic GmERD15c gene under salt stress
| 通路ID Pathway ID | KEGG通路 KEGG pathway | 1级通路 Level 1 | 2级通路 Level 2 | 上调数量 Up_number | 下调数量 Down_number | 差异数量 DEG_number | 总数量 Total_number |
|---|---|---|---|---|---|---|---|
| gmx00910 | 氮代谢 | 新陈代谢 | 能量代谢 | 6 | 2 | 8 | 44 |
| gmx00260 | 甘氨酸、丝氨酸和苏氨酸代谢 | 新陈代谢 | 氨基酸代谢 | 4 | 4 | 8 | 89 |
| gmx00940 | 苯丙烷类生物合成 | 新陈代谢 | 其他次生代谢产物的生物合成 | 8 | 7 | 15 | 260 |
| gmx00565 | 乙醚脂质代谢 | 新陈代谢 | 脂类代谢 | 5 | 0 | 5 | 43 |
| gmx00904 | 二萜类生物合成 | 新陈代谢 | 萜类化合物和聚酮类化合物的代谢 | 1 | 4 | 5 | 45 |
| gmx00564 | 甘油磷脂代谢 | 新陈代谢 | 脂类代谢 | 9 | 1 | 10 | 164 |
| gmx00592 | 亚麻酸代谢 | 新陈代谢 | 脂类代谢 | 4 | 2 | 6 | 72 |
| gmx00908 | 玉米素生物合成 | 新陈代谢 | 萜类化合物和聚酮类化合物的代谢 | 3 | 1 | 4 | 36 |
| gmx00350 | 酪氨酸代谢 | 新陈代谢 | 氨基酸代谢 | 3 | 2 | 5 | 56 |
| gmx00270 | 半胱氨酸和蛋氨酸代谢 | 新陈代谢 | 氨基酸代谢 | 6 | 3 | 9 | 154 |
| gmx00630 | 乙醛酸盐和二羧酸代谢 | 新陈代谢 | 碳水化合物代谢 | 0 | 6 | 6 | 87 |
| gmx00130 | 泛醌和其他萜类醌生物合成 | 新陈代谢 | 辅因子和维生素的代谢 | 4 | 1 | 5 | 68 |
| gmx00943 | 异黄酮生物合成 | 新陈代谢 | 其他次生代谢产物的生物合成 | 3 | 0 | 3 | 28 |
| gmx00941 | 类黄酮生物合成 | 新陈代谢 | 其他次生代谢产物的生物合成 | 3 | 2 | 5 | 77 |
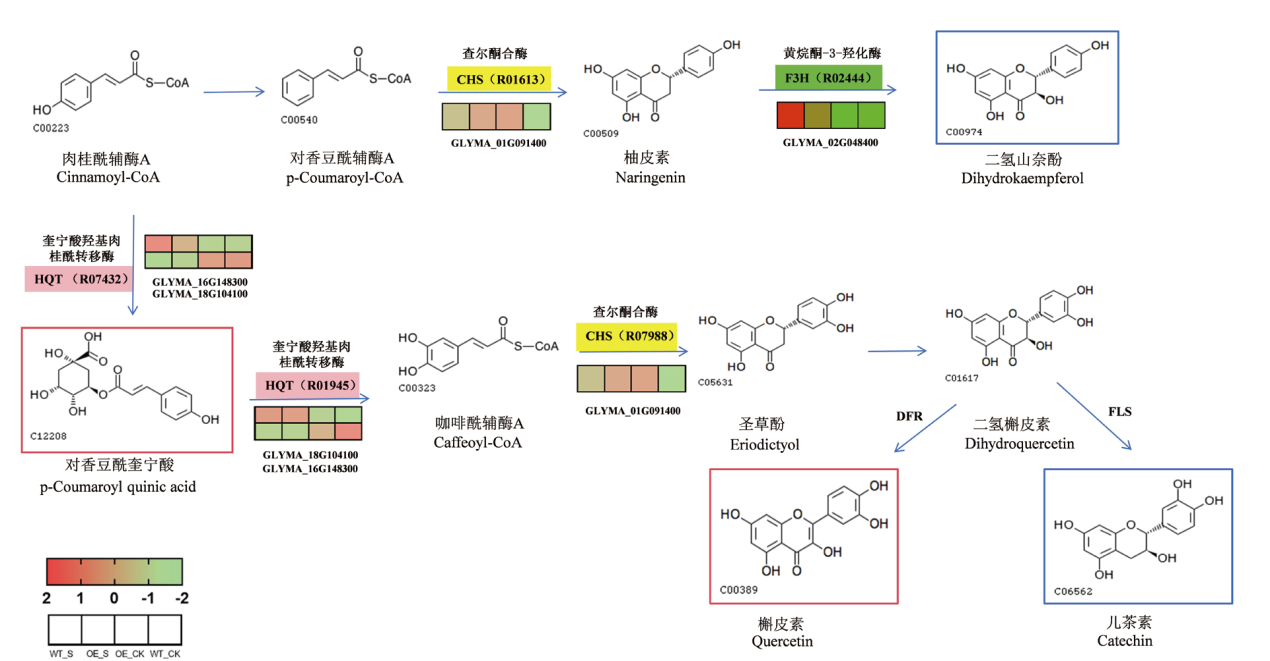
Fig. 5 Transcriptional metabolism combined with KEGG analysis The base color indicates mRNA and the rectangle indicates metabolites. Blue indicates metabolite downregulation, red indicates metabolite upregulation, green indicates mRNA downregulation, yellow indicates mRNA upregulation, and pink indicates polydifferential mRNA.The numbers in the parentheses indicate the types of enzyme reactions. R07432: Quinate hydroxycinnamoyltransferase.R01945: Quinate ester O-(3,4-dihydroxycinnamoyl)transferase. R01613: 4-coumaroyl-CoA:malate acyltransferase. R07988: Caffeoyl-CoA:malate acyltransferase. R02444: Oxidoreductase
| [1] | 王文月, 姚志鹏, 于洋, 等. 我国大豆种业科技创新发展现状及对策建议[J]. 中国农业科技导报, 2024, 26(3): 1-6. |
| Wang WY, Yao ZP, Yu Y, et al. Scientific and technological innovation of soybean seed industry in China: current situation and strategy[J]. J Agric Sci Technol, 2024, 26(3): 1-6. | |
| [2] |
Kumar A, Singh S, Gaurav AK, et al. Plant growth-promoting bacteria: biological tools for the mitigation of salinity stress in plants[J]. Front Microbiol, 2020, 11: 1216.
doi: 10.3389/fmicb.2020.01216 pmid: 32733391 |
| [3] |
Liang XY, Li JF, Yang YQ, et al. Designing salt stress-resilient crops: current progress and future challenges[J]. J Integr Plant Biol, 2024, 66(3): 303-329.
doi: 10.1111/jipb.13599 |
| [4] | Ahammed GJ, Yang YX. Anthocyanin-mediated arsenic tolerance in plants[J]. Environ Pollut, 2022, 292(Pt B): 118475. |
| [5] | Clayton WA, Albert NW, Thrimawithana AH, et al. UVR8-mediated induction of flavonoid biosynthesis for UVB tolerance is conserved between the liverwort Marchantia polymorpha and flowering plants[J]. Plant J, 2018, 96(3): 503-517. |
| [6] | Yu WW, Liu HM, Luo JQ, et al. Partial root-zone simulated drought induces greater flavonoid accumulation than full root-zone simulated water deficiency in the leaves of Ginkgo biloba[J]. Environ Exp Bot, 2022, 201: 104998. |
| [7] | Li Q, Yu HM, Meng XF, et al. Ectopic expression of glycosyltransferase UGT76E11 increases flavonoid accumulation and enhances abiotic stress tolerance in Arabidopsis[J]. Plant Biol, 2018, 20(1): 10-19. |
| [8] | Li BZ, Fan RN, Guo SY, et al. The Arabidopsis MYB transcription factor, MYB111 modulates salt responses by regulating flavonoid biosynthesis[J]. Environ Exp Bot, 2019, 166: 103807. |
| [9] | Bian XH, Li W, Niu CF, et al. A class B heat shock factor selected for during soybean domestication contributes to salt tolerance by promoting flavonoid biosynthesis[J]. New Phytol, 2020, 225(1): 268-283. |
| [10] | Aalto MK, Helenius E, Kariola T, et al. ERD15—an attenuator of plant ABA responses and stomatal aperture[J]. Plant Sci, 2012, 182: 19-28. |
| [11] | Wu GF, Tian NF, She FW, et al. Characteristics analysis of Early Responsive to Dehydration genes in Arabidopsis thaliana(AtER-D)[J]. Plant Signal Behav, 2023, 18(1): 2105021. |
| [12] | Huang YM, Du BS, Yu MX, et al. Picea wilsonii NAC31 and DREB2A cooperatively activate ERD1 to modulate drought resistance in transgenic Arabidopsis[J]. Int J Mol Sci, 2024, 25(4): 2037. |
| [13] |
Kariola T, Brader G, Helenius E, et al. EARLY RESPONSIVE TO DEHYDRATION 15, a negative regulator of abscisic acid responses in Arabidopsis[J]. Plant Physiol, 2006, 142(4): 1559-1573.
doi: 10.1104/pp.106.086223 pmid: 17056758 |
| [14] |
Jin T, Sun YY, Shan Z, et al. Natural variation in the promoter of GsERD15B affects salt tolerance in soybean[J]. Plant Biotechnol J, 2021, 19(6): 1155-1169.
doi: 10.1111/pbi.13536 pmid: 33368860 |
| [15] | Slawinski L, Israel A, Artault C, et al. Responsiveness of early response to dehydration six-like transporter genes to water deficit in Arabidopsis thaliana leaves[J]. Front Plant Sci, 2021, 12: 708876. |
| [16] |
Farrant JM, Cooper K, Hilgart A, et al. A molecular physiological review of vegetative desiccation tolerance in the resurrection plant Xerophyta viscosa(Baker)[J]. Planta, 2015, 242(2): 407-426.
doi: 10.1007/s00425-015-2320-6 pmid: 25998524 |
| [17] | Ziaf K, Loukehaich R, Gong PJ, et al. A multiple stress-responsive gene ERD15 from Solanum pennellii confers stress tolerance in tobacco[J]. Plant Cell Physiol, 2011, 52(6): 1055-1067. |
| [18] | 曹沙沙. 大豆GmERD15c基因的克隆及其功能分析[D]. 长春: 吉林农业大学, 2022. |
| Cao SS. Cloning and functional analysis of GmERD15c gene in soybean[D]. Changchun: Jilin Agricultural University, 2022. | |
| [19] |
Wu JT, Lv SD, Zhao L, et al. Advances in the study of the function and mechanism of the action of flavonoids in plants under environmental stresses[J]. Planta, 2023, 257(6): 108.
doi: 10.1007/s00425-023-04136-w pmid: 37133783 |
| [20] | Li SC. Novel insight into functions of ascorbate peroxidase in higher plants: more than a simple antioxidant enzyme[J]. Redox Biol, 2023, 64: 102789. |
| [21] | Speisky H, Shahidi F, Costa de Camargo A, et al. Revisiting the oxidation of flavonoids: loss, conservation or enhancement of their antioxidant properties[J]. Antioxidants, 2022, 11(1): 133. |
| [22] | Wang ZL, Wang S, Kuang Y, et al. A comprehensive review on phytochemistry, pharmacology, and flavonoid biosynthesis of Scutellaria baicalensis[J]. Pharm Biol, 2018, 56(1): 465-484. |
| [23] | Dong NQ, Lin HX. Contribution of phenylpropanoid metabolism to plant development and plant-environment interactions[J]. J Integr Plant Biol, 2021, 63(1): 180-209. |
| [24] | Uleberg E, Rohloff J, Jaakola L, et al. Effects of temperature and photoperiod on yield and chemical composition of northern and southern clones of bilberry(Vaccinium myrtillus L.)[J]. J Agric Food Chem, 2012, 60(42): 10406-10414. |
| [25] |
Niggeweg R, Michael AJ, Martin C. Engineering plants with increased levels of the antioxidant chlorogenic acid[J]. Nat Biotechnol, 2004, 22(6): 746-754.
doi: 10.1038/nbt966 pmid: 15107863 |
| [26] | Guo J, Carrington Y, Alber A, et al. Molecular characterization of quinate and shikimate metabolism in Populus trichocarpa[J]. J Biol Chem, 2014, 289(34): 23846-23858. |
| [27] | Clé C, Hill LM, Niggeweg R, et al. Modulation of chlorogenic acid biosynthesis in Solanum lycopersicum; consequences for phenolic accumulation and UV-tolerance[J]. Phytochemistry, 2008, 69(11): 2149-2156. |
| [28] | Deng XB, Bashandy H, Ainasoja M, et al. Functional diversification of duplicated chalcone synthase genes in anthocyanin biosynthesis of Gerbera hybrida[J]. New Phytol, 2014, 201(4): 1469-1483. |
| [29] | Zhang XB, Abrahan C, Colquhoun TA, et al. A proteolytic regulator controlling Chalcone synthase stability and flavonoid biosynthesis in Arabidopsis[J]. Plant Cell, 2017, 29(5): 1157-1174. |
| [30] |
Schijlen EGWM, de Vos CH, Martens S, et al. RNA interference silencing of chalcone synthase, the first step in the flavonoid biosynthesis pathway, leads to parthenocarpic tomato fruits[J]. Plant Physiol, 2007, 144(3): 1520-1530.
doi: 10.1104/pp.107.100305 pmid: 17478633 |
| [31] | Song XY, Diao JJ, Ji J, et al. Molecular cloning and identification of a flavanone 3-hydroxylase gene from Lycium chinense, and its overexpression enhances drought stress in tobacco[J]. Plant Physiol Biochem, 2016, 98: 89-100. |
| [32] |
Tu YH, Liu F, Guo DD, et al. Molecular characterization of flavanone 3-hydroxylase gene and flavonoid accumulation in two chemotyped safflower lines in response to methyl jasmonate stimulation[J]. BMC Plant Biol, 2016, 16(1): 132.
doi: 10.1186/s12870-016-0813-5 pmid: 27286810 |
| [33] | Busche M, Acatay C, Martens S, et al. Functional characterisation of banana(Musa spp.) 2-oxoglutarate-dependent dioxygenases involved in flavonoid biosynthesis[J]. Front Plant Sci, 2021, 12: 701780. |
| [34] | Wang LX, Lui ACW, Lam PY, et al. Transgenic expression of flavanone 3-hydroxylase redirects flavonoid biosynthesis and alleviates anthracnose susceptibility in sorghum[J]. Plant Biotechnol J, 2020, 18(11): 2170-2172. |
| [35] | Liu W, Feng Y, Yu S, et al. The flavonoid biosynthesis network in plants[J]. Int J Mol Sci, 2021, 22(23): 12824. |
| [36] | Mou JL, Zhang ZH, Qiu HJ, et al. Multiomics-based dissection of Citrus flavonoid metabolism using a Citrus reticulata × Poncirus trifoliata population[J]. Hortic Res, 2021, 8(1): 56. |
| [37] | Meng XY, Li YQ, Zhou TT, et al. Functional differentiation of duplicated flavonoid 3- O-glycosyltransferases in the flavonol and anthocyanin biosynthesis of Freesia hybrida[J]. Front Plant Sci, 2019, 10: 1330. |
| [38] | Singh P, Arif Y, Bajguz A, et al. The role of quercetin in plants[J]. Plant Physiol Biochem, 2021, 166: 10-19. |
| [39] |
Martens S, Preuss A, Matern U. Multifunctional flavonoid dioxygenases: flavonol and anthocyanin biosynthesis in Arabidopsis thaliana L[J]. Phytochemistry, 2010, 71(10): 1040-1049.
doi: 10.1016/j.phytochem.2010.04.016 pmid: 20457455 |
| [40] | Yan HL, Pei XN, Zhang H, et al. MYB-mediated regulation of anthocyanin biosynthesis[J]. Int J Mol Sci, 2021, 22(6): 3103. |
| [41] | Hinojosa-Gómez J, San Martín-Hernández C, Heredia JB, et al. Anthocyanin induction by drought stress in the Calyx of Roselle cultivars[J]. Molecules, 2020, 25(7): 1555. |
| [42] | Chen WF, Xiao ZC, Wang YL, et al. Competition between anthocyanin and kaempferol glycosides biosynthesis affects pollen tube growth and seed set of Malus[J]. Hortic Res, 2021, 8(1): 173. |
| [43] |
Yan JH, Wang B, Zhong YP, et al. The soybean R2R3 MYB transcription factor GmMYB100 negatively regulates plant flavonoid biosynthesis[J]. Plant Mol Biol, 2015, 89(1-2): 35-48.
doi: 10.1007/s11103-015-0349-3 pmid: 26231207 |
| [44] | Wang X, Dai WW, Liu C, et al. Evaluation of physiological coping strategies and quality substances in purple SweetPotato under different salinity levels[J]. Genes, 2022, 13(8): 1350. |
| [45] | Xu ZC, Wang M, Ren TT, et al. Comparative transcriptome analysis reveals the molecular mechanism of salt tolerance in Apocynum venetum[J]. Plant Physiol Biochem, 2021, 167: 816-830. |
| [1] | WANG Rui, QI Ji. Integrating Histological Image Information to Enhance Cell Clustering Resolution in Spatial Transcriptome [J]. Biotechnology Bulletin, 2024, 40(8): 39-46. |
| [2] | WU Shuai, XIN Yan-ni, MAI Chun-hai, MU Xiao-ya, WANG Min, YUE Ai-qin, ZHAO Jin-zhong, WU Shen-jie, DU Wei-jun, WANG Li-xiang. Genome-wide Identification and Stress Response Analysis of Soybean GS Gene Family [J]. Biotechnology Bulletin, 2024, 40(8): 63-73. |
| [3] | GAO Meng-meng, ZHAO Tian-yu, JIAO Xin-yue, LIN Chun-jing, GUAN Zhe-yun, DING Xiao-yang, SUN Yan-yan, ZHANG Chun-bao. Comparative Transcriptome Analysis of Cytoplasmic Male Sterile Line and Its Restorer Line in Soybean [J]. Biotechnology Bulletin, 2024, 40(7): 137-149. |
| [4] | WANG Fang, YU Lu, QI Ze-zheng, ZHOU Chang-jun, YU Ji-dong. Screening and Biocontrol Effect of Antagonistic Bacteria against Soybean Root Rot [J]. Biotechnology Bulletin, 2024, 40(7): 216-225. |
| [5] | BAI Zhi-yuan, XU Fei, YANG Wu, WANG Ming-gui, YANG Yu-hua, ZHANG Hai-ping, ZHANG Rui-jun. Transcriptome Analysis of Fertility Transformation in Weakly Restoring Hybrid F1 of Soybean Cytoplasmic Male Sterility [J]. Biotechnology Bulletin, 2024, 40(6): 134-142. |
| [6] | LOU Yin, GAO Hao-jun, WANG Xi, NIU Jing-ping, WANG Min, DU Wei-jun, YUE Ai-qin. Identification and Expression Pattern Analysis of GmHMGS Gene in Soybean [J]. Biotechnology Bulletin, 2024, 40(4): 110-121. |
| [7] | LI Can, JIANG Xiang-ning, GAI Ying. Cloning of the LkF3H2 Gene in Larix kaempferi and Its Function in Regulating Flavonoid Metabolism [J]. Biotechnology Bulletin, 2024, 40(2): 245-252. |
| [8] | XU Pei-dong, YI Jian-feng, CHEN Di, CHEN Hao, XIE Bing-yan, ZHAO Wen-jun. Progress in the Application of Omics Technology in Biocontrol Bacillus [J]. Biotechnology Bulletin, 2024, 40(10): 208-220. |
| [9] | JIANG Yu-shan, LAN Qian, WANG Fang, JIANG Liang, PEI Cheng-cheng. Characterization of a Quinoa Mutant Affecting Tyrosine Metabolism [J]. Biotechnology Bulletin, 2024, 40(10): 253-261. |
| [10] | ZHAO Rui, DI Jing-yi, ZHANG Guang-tong, LIU Hao, GAO Wei-xia. Screening Endogenous Expression Elements in Streptococcus zooepidemicus via Transcriptomics Analysis and Applications for High Yield of Hyaluronic Acid [J]. Biotechnology Bulletin, 2024, 40(10): 296-304. |
| [11] | HE Shi-yu, ZENG Zhong-da, LI Bo-yan. Application Progress of Spatially Resolved Metabolomics in Disease Diagnosis Research [J]. Biotechnology Bulletin, 2024, 40(1): 145-159. |
| [12] | ZHOU Ai-ting, PENG Rui-qi, WANG Fang, WU Jian-rong, MA Huan-cheng. Analysis of Metabolic Differences of Biocontrol Strain DZY6715 at Different Growth Stages [J]. Biotechnology Bulletin, 2023, 39(9): 225-235. |
| [13] | WANG Shuai, FENG Yu-mei, BAI Miao, DU Wei-jun, YUE Ai-qin. Functional Analysis of Soybean Gene GmHMGR Responding to Exogenous Hormones and Abiotic Stresses [J]. Biotechnology Bulletin, 2023, 39(7): 131-142. |
| [14] | LI Wen-chen, LIU Xin, KANG Yue, LI Wei, QI Ze-zheng, YU Lu, WANG Fang. Optimization and Application of Tobacco Rattle Virus-induced Gene Silencing System in Soybean [J]. Biotechnology Bulletin, 2023, 39(7): 143-150. |
| [15] | HAN Hua-rui, YANG Yu-lu, MEN Yi-han, HAN Shang-ling, HAN Yuan-huai, HUO Yi-qiong, HOU Si-yu. SiYABBYs Involved in Rhamnoside Biosynthesis During the Flower Development of Setaria italica, Based on Metabolomics [J]. Biotechnology Bulletin, 2023, 39(6): 189-198. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||