








Biotechnology Bulletin ›› 2024, Vol. 40 ›› Issue (6): 190-202.doi: 10.13560/j.cnki.biotech.bull.1985.2024-0012
Previous Articles Next Articles
LI Bo-jing( ), ZHENG La-mei, WU Wu-yun, GAO Fei(
), ZHENG La-mei, WU Wu-yun, GAO Fei( ), ZHOU Yi-jun(
), ZHOU Yi-jun( )
)
Received:2024-01-04
Online:2024-06-26
Published:2024-06-24
Contact:
GAO Fei, ZHOU Yi-jun
E-mail:c2236363710@163.com;gaofei@muc.edu.cn;zhouyijun@muc.edu.cn
LI Bo-jing, ZHENG La-mei, WU Wu-yun, GAO Fei, ZHOU Yi-jun. Evolution, Expression, and Functional Analysis of the HSP20 Gene Family from Simmondisa chinensis[J]. Biotechnology Bulletin, 2024, 40(6): 190-202.
| 基因名称Gene name | 上游引物 Forward primer(5'-3') | 下游引物 Reverse primer(5'-3') |
|---|---|---|
| 18S | cgttaacgaacgagacctca | cccagaacatctaagggcat |
| ScHSP20-1 | gttgcggtcgcatttgttg | cgtccacggaagccatagaa |
| ScHSP20-3 | gccctgaaaatgaagagggtac | cctttgaacttggcttggatg |
| ScHSP20-4 | aatcaaaggagaaggggagaaa | aggaaccaccaccttcaacact |
| ScHSP20-13 | cgtcaagagggactcaccaaa | caccagaaaccgaaggagga |
| ScHSP20-14 | tcaagagggactcaccaaacag | agccacgtaagccccaatc |
| ScHSP20-16 | acacccttcatcacatcctcg | agctgttggggtactccttcac |
| ScHSP20-17 | accgtgtagagcggagcagt | cagtgacagtcaggaccccat |
| ScHSP20-20 | cagtaagagggcggaggatg | aagggaaagacgacgaggc |
| ScHSP20-25 | atggcgagcaaggcgttgacatgca | tgcatgtcaacgccttgctcgccat |
| ScHSP20-28 | tggcctctgtacgtgctgatt | caccctgttctcctccacttct |
| ScHSP20-23 | tgaggaacctccgacaaagac | cctcctgatgttgacgatgaaa |
| ScHSP20-26 | tccaggtcttagcaaggagca | tgtctttccctgtgggttcg |
| ScHSP20-32 | ctccctctgactgggtttcg | gggcgttctccacttatcctta |
Table 1 Primers used in RT-qPCR
| 基因名称Gene name | 上游引物 Forward primer(5'-3') | 下游引物 Reverse primer(5'-3') |
|---|---|---|
| 18S | cgttaacgaacgagacctca | cccagaacatctaagggcat |
| ScHSP20-1 | gttgcggtcgcatttgttg | cgtccacggaagccatagaa |
| ScHSP20-3 | gccctgaaaatgaagagggtac | cctttgaacttggcttggatg |
| ScHSP20-4 | aatcaaaggagaaggggagaaa | aggaaccaccaccttcaacact |
| ScHSP20-13 | cgtcaagagggactcaccaaa | caccagaaaccgaaggagga |
| ScHSP20-14 | tcaagagggactcaccaaacag | agccacgtaagccccaatc |
| ScHSP20-16 | acacccttcatcacatcctcg | agctgttggggtactccttcac |
| ScHSP20-17 | accgtgtagagcggagcagt | cagtgacagtcaggaccccat |
| ScHSP20-20 | cagtaagagggcggaggatg | aagggaaagacgacgaggc |
| ScHSP20-25 | atggcgagcaaggcgttgacatgca | tgcatgtcaacgccttgctcgccat |
| ScHSP20-28 | tggcctctgtacgtgctgatt | caccctgttctcctccacttct |
| ScHSP20-23 | tgaggaacctccgacaaagac | cctcctgatgttgacgatgaaa |
| ScHSP20-26 | tccaggtcttagcaaggagca | tgtctttccctgtgggttcg |
| ScHSP20-32 | ctccctctgactgggtttcg | gggcgttctccacttatcctta |
| 用途 Purpose | 基因名称 Gene name | 上游引物 Forward primer(5'-3') | 下游引物 Reverse primer(5'-3') |
|---|---|---|---|
| 原核表达载体 Prokaryotic expression vector | ScHSP20-17 | cgcgtggatccccggATGTCGCTTATTCCAAGTAGGT | agtcagtcacgatgcACCAGAGATTTCAATGGCTTTG |
| 酵母表达载体 Yeast expression vector | ScHSP20-17 | ctatagggaatattaATGTCGCTTATTCCAAGTAGGT | cggccgttactagtgACCAGAGATTTCAATGGCTTTG |
| 亚细胞定位载体 Subcellular localization vector | ScHSP20-17 | agtccggagctagctATGTCGCTTATTCCAAGTAGGT | cccttgctcaccatgACCAGAGATTTCAATGGCTTTG |
| 内参基因 Internal reference gene | Actin3 | GAGGGCCGTGTTCCCCAGCATCGTC | TCTTTTTGATTGAGCCTCATCCCCT |
| 基因表达分析 Gene expression analysis | ScHSP20-17 | AAGAATGATACCTGGCACCG | TGACCTCCTCCTTAGGCACA |
Table 2 Primer sequences
| 用途 Purpose | 基因名称 Gene name | 上游引物 Forward primer(5'-3') | 下游引物 Reverse primer(5'-3') |
|---|---|---|---|
| 原核表达载体 Prokaryotic expression vector | ScHSP20-17 | cgcgtggatccccggATGTCGCTTATTCCAAGTAGGT | agtcagtcacgatgcACCAGAGATTTCAATGGCTTTG |
| 酵母表达载体 Yeast expression vector | ScHSP20-17 | ctatagggaatattaATGTCGCTTATTCCAAGTAGGT | cggccgttactagtgACCAGAGATTTCAATGGCTTTG |
| 亚细胞定位载体 Subcellular localization vector | ScHSP20-17 | agtccggagctagctATGTCGCTTATTCCAAGTAGGT | cccttgctcaccatgACCAGAGATTTCAATGGCTTTG |
| 内参基因 Internal reference gene | Actin3 | GAGGGCCGTGTTCCCCAGCATCGTC | TCTTTTTGATTGAGCCTCATCCCCT |
| 基因表达分析 Gene expression analysis | ScHSP20-17 | AAGAATGATACCTGGCACCG | TGACCTCCTCCTTAGGCACA |
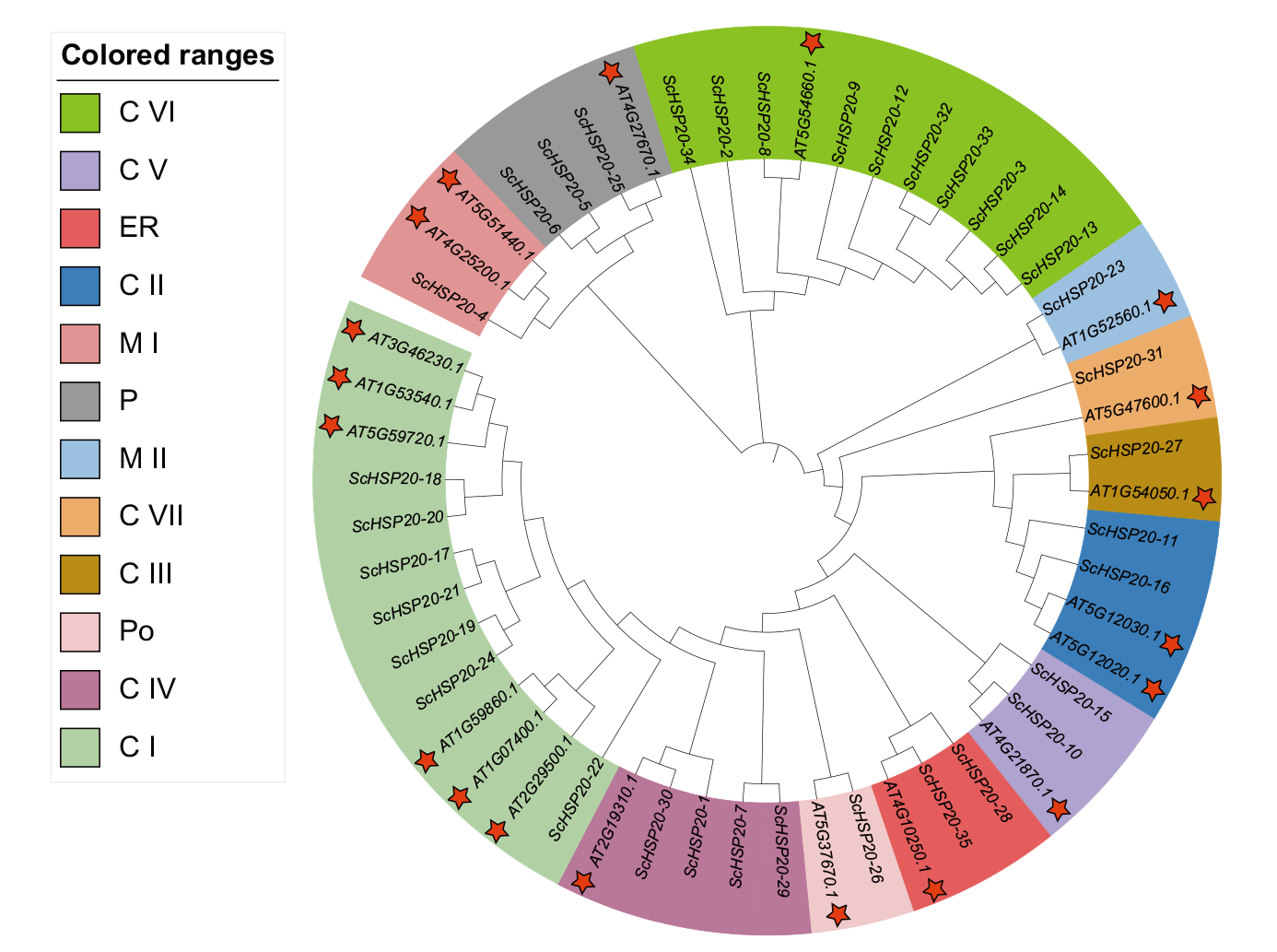
Fig. 3 Evolutionary analysis of the HSP20 gene in Arabidopsis and S. chinensis C I, C II, C III, CIV, C V, C VI, C VII: Cytoplasm or nucleus. Ml, MII: Mitochondria. ER: Endoplasmic reticulum. P: Plastid. Po: Peroxisome
| 重复基因对 Repeated gene pairs | Ka | Ks | Ka/Ks |
|---|---|---|---|
| ScHSP20-2 vs ScHSP20-17 | 0.383 89 | 2.622 38 | 0.146 39 |
| ScHSP20-5 vs ScHSP20-6 | 0.003 77 | 0.027 34 | 0.137 86 |
| ScHSP20-17 vs ScHSP20-24 | 0.137 24 | 1.495 32 | 0.091 78 |
Table 3 Ka/Ks values of fragment repeat gene pairs in ScHSP20 family members
| 重复基因对 Repeated gene pairs | Ka | Ks | Ka/Ks |
|---|---|---|---|
| ScHSP20-2 vs ScHSP20-17 | 0.383 89 | 2.622 38 | 0.146 39 |
| ScHSP20-5 vs ScHSP20-6 | 0.003 77 | 0.027 34 | 0.137 86 |
| ScHSP20-17 vs ScHSP20-24 | 0.137 24 | 1.495 32 | 0.091 78 |
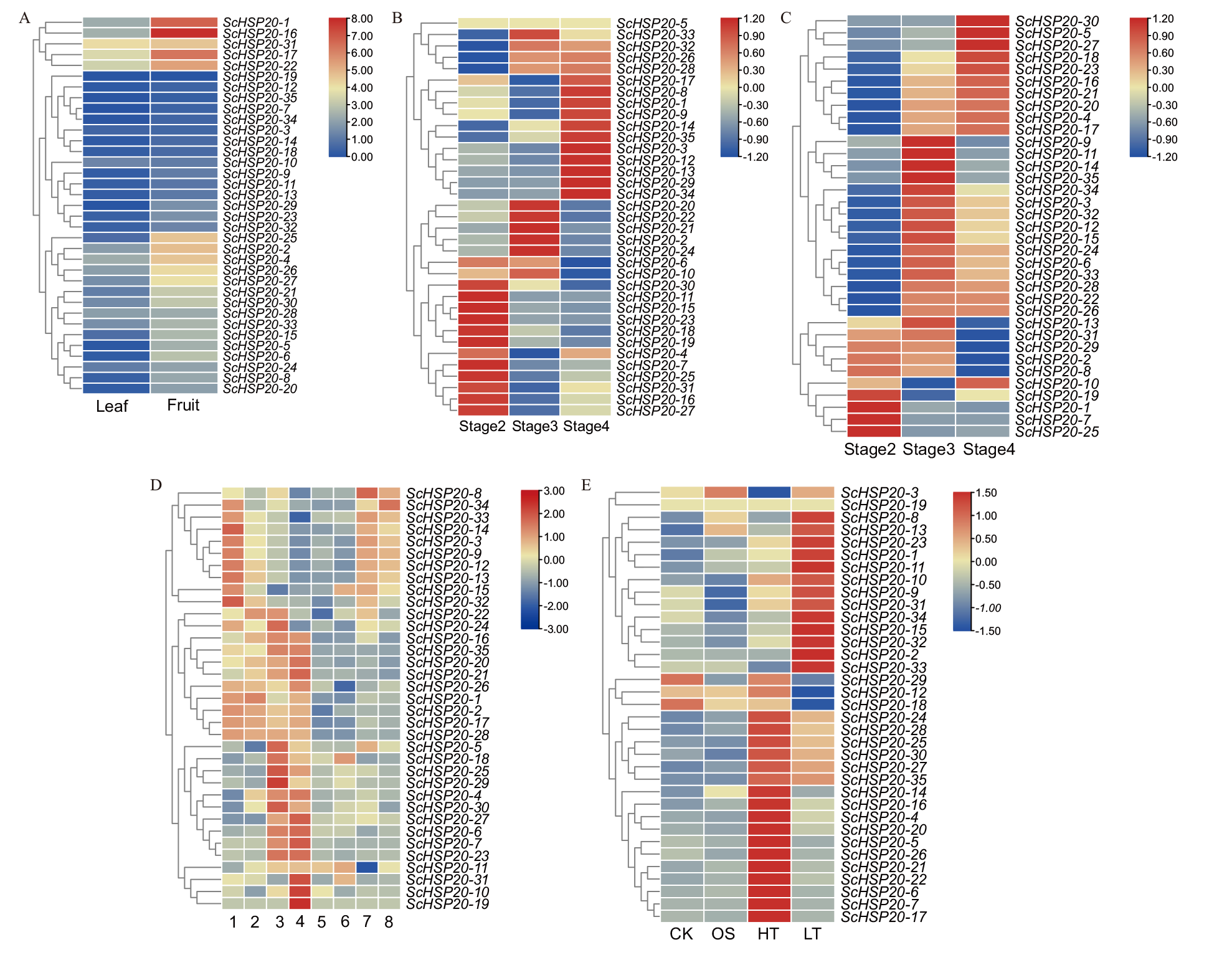
Fig. 5 Expressions of ScHSP20 gene in different tissues and under different stresses A: Leaves and fruits of Simondwood. B: Male flowers at different flowering stages. C: Different flowering stages of female flowers in Simondmu. D: Different stages and parts of seed development in Simondwood(1: Early developmental seeds. 2: Mid stage developmental seeds. 3: Late developmental seeds. 4: Dry seeds. 5: Middle stage development of seed cotyledons. 6: Mid developmental seed hypocotyl. 7: Mid stage development of seed coat). E: The expression of ScHSP20 gene under osmotic(OS), high temperature(HT), and low temperature(LT)stress
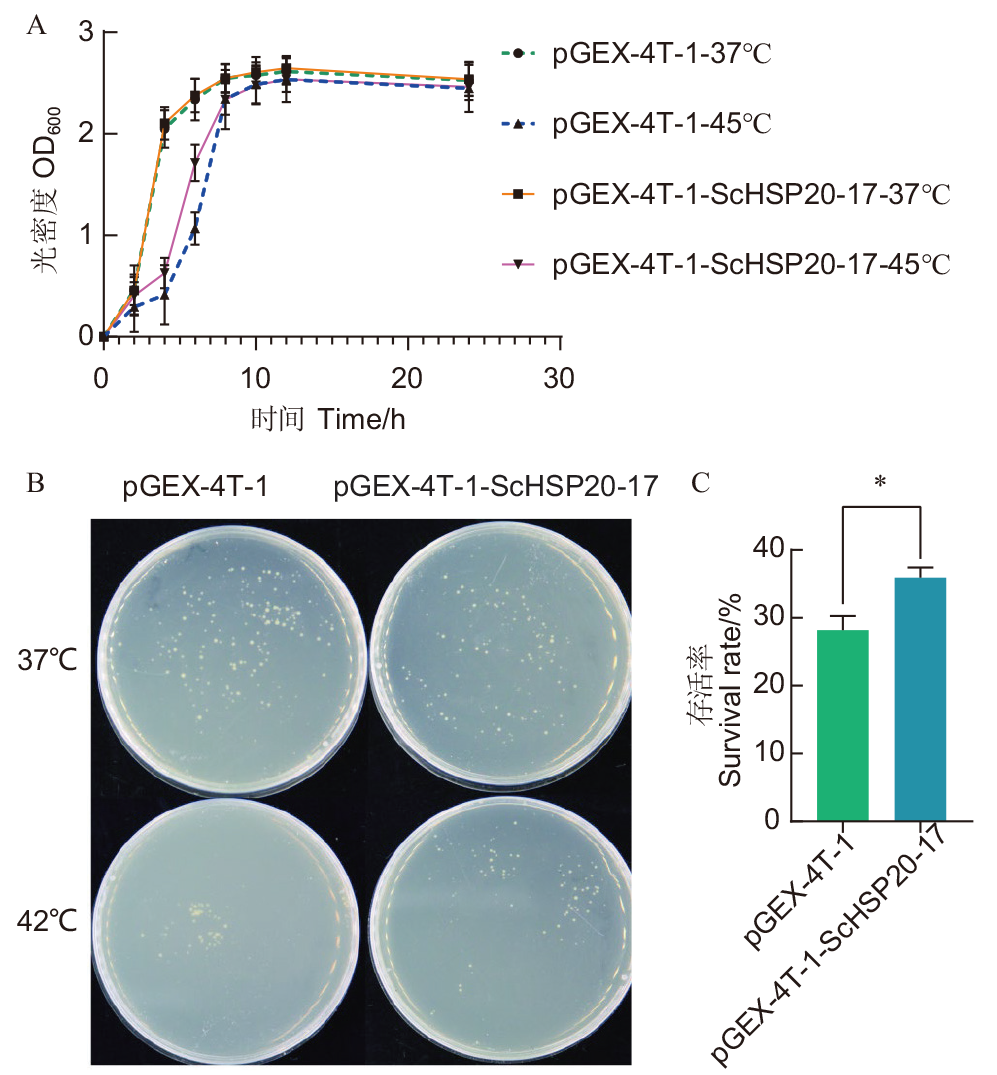
Fig. 9 Effects of heterologous expression of ScHSP20-17 gene on the tolerance of Escherichia coli to high temperature stress A: rowth curves of recombinant bacteria and control bacteria under high temperature stress. B: Growth of control and recombinant bacteria before and after treatment.C: Statistics of survival rate of control bacteria and recombinant bacteria before and after high temperature stress treatment. * indicate significant differences at P<0.05

Fig. 11 Effects of heterologous expression of ScHSP20-17 gene on the tolerance of tobacco to high temperature stress A: Image of tobacco without injection of bacterial liquid under fluorescent protein lamp irradiation. B: Image of tobacco with injection of bacterial liquid under fluorescent protein lamp irradiation. C: Wild type tobacco(WT), tobacco with injection of empty bacterial liquid(PRI), and tobacco leaves with injection of bacterial liquid carrying PRI-ScHSP20-17-GFP vector qRT PCR analysis. D-E: Wild type tobacco. F: Tobacco with instantaneous rotation of empty liquid(PRI). G: Tobacco with instantaneous rotation of ScHSP20-17. H: MDA content determination
| [1] | Zheng LM, Wu WY, Gao YF, et al. Integrated transcriptome, small RNA and degradome analysis provide insights into the transcriptional regulatory networks underlying cold acclimation in jojoba[J]. Sci Hortic, 2022, 299. |
| [2] | Kawiński A, Miklaszewska M, Stelter S, et al. Lipases of germinating jojoba seeds efficiently hydrolyze triacylglycerols and wax esters and display wax ester-synthesizing activity[J]. BMC Plant Biol, 2021, 21(1): 50. |
| [3] | Gad HA, Roberts A, Hamzi SH, et al. Jojoba oil: An updated comprehensive review on chemistry, pharmaceutical uses, and toxicity[J]. Polymers, 2021, 13(11): 1711. |
| [4] | Pazyar N, Yaghoobi R, Ghassemi MR, et al. Jojoba in dermatology: a succinct review[J]. G Ital Dermatol Venereol., 2013, 148(6): 687-691. |
| [5] | Zhu JK. Abiotic stress signaling and responses in plants[J]. Cell, 2016, 167(2): 313-324. |
| [6] |
Neta-Sharir I, Isaacson T, Lurie S, et al. Dual role for tomato heat shock protein 21: protecting photosystem II from oxidative stress and promoting color changes during fruit maturation[J]. Plant Cell, 2005, 17(6): 1829-1838.
doi: 10.1105/tpc.105.031914 pmid: 15879560 |
| [7] | Yu JH, Cheng Y, Feng K, et al. Genome-wide identification and expression profiling of tomato HSP20 gene family in response to biotic and abiotic stresses[J]. Front Plant Sci, 2016, 7: 1215. |
| [8] |
Wang WX, Vinocur B, Shoseyov O, et al. Role of plant heat-shock proteins and molecular chaperones in the abiotic stress response[J]. Trends Plant Sci, 2004, 9(5): 244-252.
doi: 10.1016/j.tplants.2004.03.006 pmid: 15130550 |
| [9] |
Waters ER. The evolution, function, structure, and expression of the plant sHSPs[J]. J Exp Bot, 2013, 64(2): 391-403.
doi: 10.1093/jxb/ers355 pmid: 23255280 |
| [10] | Qi XY, Di ZX, Li YY, et al. Genome-wide identification and expression profiling of heat shock protein 20 gene family in Sorbus pohuashanensis(Hance)hedl under abiotic stress[J]. Genes, 2022, 13(12): 2241. |
| [11] |
Jaya N, Garcia V, Vierling E. Substrate binding site flexibility of the small heat shock protein molecular chaperones[J]. Proc Natl Acad Sci USA, 2009, 106(37): 15604-15609.
doi: 10.1073/pnas.0902177106 pmid: 19717454 |
| [12] |
Sun WN, Van Montagu M, Verbruggen N. Small heat shock proteins and stress tolerance in plants[J]. Biochim Biophys Acta, 2002, 1577(1): 1-9.
doi: 10.1016/s0167-4781(02)00417-7 pmid: 12151089 |
| [13] |
Haslbeck M, Vierling E. A first line of stress defense: small heat shock proteins and their function in protein homeostasis[J]. J Mol Biol, 2015, 427(7): 1537-1548.
doi: 10.1016/j.jmb.2015.02.002 pmid: 25681016 |
| [14] | Ji XR, Yu YH, Ni PY, et al. Genome-wide identification of small heat-shock protein(HSP20)gene family in grape and expression profile during berry development[J]. BMC Plant Biol, 2019, 19(1): 433. |
| [15] |
Ouyang YD, Chen JJ, Xie WB, et al. Comprehensive sequence and expression profile analysis of HSP20 gene family in rice[J]. Plant Mol Biol, 2009, 70(3): 341-357.
doi: 10.1007/s11103-009-9477-y pmid: 19277876 |
| [16] |
Lopes-Caitar VS, Darben LM, et al. Genome-wide analysis of the HSP20 gene family in soybean: comprehensive sequence, genomic organization and expression profile analysis under abiotic and biotic stresses[J]. BMC Genomics, 2013, 14: 577.
doi: 10.1186/1471-2164-14-577 pmid: 23985061 |
| [17] |
Guo M, Liu JH, Lu JP, et al. Genome-wide analysis of the CaHsp20 gene family in pepper: comprehensive sequence and expression profile analysis under heat stress[J]. Front Plant Sci, 2015, 6: 806.
doi: 10.3389/fpls.2015.00806 pmid: 26483820 |
| [18] | Scharf KD, Siddique M, Vierling E. The expanding family of Arabidopsis thaliana small heat stress proteins and a new family of proteins containing alpha-crystallin domains(Acd proteins)[J]. Cell Stress and Chaperones, 2001, 6(3): 225-237. |
| [19] |
Chen CJ, Chen H, Zhang Y, et al. TBtools: An integrative toolkit developed for interactive analyses of big biological data[J]. Mol Plant, 2020, 13(8): 1194-1202.
doi: S1674-2052(20)30187-8 pmid: 32585190 |
| [20] | Sturtevant D, Lu SP, Zhou ZW, et al. The genome of jojoba(Simmondsia chinensis): A taxonomically isolated species that directs wax ester accumulation in its seeds[J]. Sci Adv, 2020, 6(11): eaay3240. |
| [21] |
孙小倩, 王佳蕊, 陈庆富, 等. 苦荞转录因子FtMYBF的克隆、亚细胞定位及表达分析[J]. 生物技术通报, 2021, 37(3): 10-17.
doi: 10.13560/j.cnki.biotech.bull.1985.2020-0861 |
| Sun XQ, Wang JR, Chen QF, et al. Cloning, subcellular localization, and expression analysis of the transcription factor FtMYBF from tartary buckwheat[J]. Biotechnol Bulletin, 2021, 37(3): 10-17. | |
| [22] | Yao FW, Song CH, Wang HT, et al. Genome-wide characterization of the HSP20 gene family identifies potential members involved in temperature stress response in apple[J]. Front Genet, 2020, 11: 609184. |
| [23] | Aghdam MS, Sevillano L, Flores FB, et al. Heat shock proteins as biochemical markers for postharvest chilling stress in fruits and vegetables[J]. Sci Hortic, 2013, 160: 54-64. |
| [24] | Hu YP, Zhang TT, Liu Y, et al. Pumpkin(Cucurbita moschata)HSP20 gene family identification and expression under heat stress[J]. Front Genet, 2021, 12: 753953. |
| [25] | Huang JJ, Hai ZX, Wang RY, et al. Genome-wide analysis of HSP20 gene family and expression patterns under heat stress in cucumber(Cucumis sativus L.)[J]. Front Plant Sci, 2022, 13: 968418. |
| [26] | Lian XD, Wang QP, Li TH, et al. Phylogenetic and transcriptional analyses of the HSP20 gene family in peach revealed that PpHSP20-32 is involved in plant height and heat tolerance[J]. Int J Mol Sci, 2022, 23(18): 10849. |
| [27] |
Nagaraju M, Reddy PS, Kumar SA, et al. Genome-wide identification and transcriptional profiling of small heat shock protein gene family under diverse abiotic stress conditions in Sorghum bicolor(L.)[J]. Int J Biol Macromol, 2020, 142: 822-834.
doi: S0141-8130(19)35485-6 pmid: 31622710 |
| [28] |
Xu GX, Guo CC, Shan HY, et al. Divergence of duplicate genes in exon-intron structure[J]. Proc Natl Acad Sci USA, 2012, 109(4): 1187-1192.
doi: 10.1073/pnas.1109047109 pmid: 22232673 |
| [29] |
Mattick JS, Gagen MJ. The evolution of controlled multitasked gene networks: the role of introns and other noncoding RNAs in the development of complex organisms[J]. Mol Biol Evol, 2001, 18(9): 1611-1130.
doi: 10.1093/oxfordjournals.molbev.a003951 pmid: 11504843 |
| [30] | Chung BYW, Simons C, Firth AE, et al. Effect of 5'UTR introns on gene expression in Arabidopsis thaliana[J]. BMC Genomics, 2006, 7: 120. |
| [31] | Sarkar NK, Kim YK, Grover A. Rice shsp genes: genomic organization and expression profiling under stress and development[J]. BMC Genom, 2009, 10(1): 393. |
| [32] |
Vision TJ, Brown DG, Tanksley SD. The origins of genomic duplications in Arabidopsis[J]. Science, 2000, 290(5499): 2114-2117.
doi: 10.1126/science.290.5499.2114 pmid: 11118139 |
| [33] |
Conant GC, Wolfe KH. Turning a hobby into a job: how duplicated genes find new functions[J]. Nat Rev Genet, 2008, 9(12): 938-950.
doi: 10.1038/nrg2482 pmid: 19015656 |
| [34] | De Grassi A, Lanave C, Saccone C. Genome duplication and gene-family evolution: the case of three OXPHOS gene families[J]. Gene, 2008, 421(1/2): 1-6. |
| [35] | Flagel LE, Wendel JF. Gene duplication and evolutionary novelty in plants[J]. The New Phytol, 2009, 183(3): 557-564. |
| [36] | Hua YG, Liu Q, Zhai YF, et al. Genome-wide analysis of the HSP20 gene family and its response to heat and drought stress in Coix(Coix lacryma-jobi L.)[J]. BMC Genomics, 2023, 24(1): 478. |
| [37] |
Yamaguchi-Shinozaki K, Shinozaki K. Organization of cis-acting regulatory elements in osmotic- and cold-stress-responsive promoters[J]. Trends Plant Sci, 2005, 10(2): 88-94.
doi: 10.1016/j.tplants.2004.12.012 pmid: 15708346 |
| [38] | Tian C, Zhang ZY, Huang Y, et al. Functional characterization of the Pinellia ternata cytoplasmic class II small heat shock protein gene PtsHSP17.2 via promoter analysis and overexpression in tobacco[J]. Plant Physiol Biochem, 2022, 177: 1-9. |
| [1] | WANG Yu-shu, ZHAO Lin-lin, ZHAO Shuang, HU Qi, BAI Hui-xia, WANG Huan, CAO Ye-ping, FAN Zhen-yu. Cloning and Expression Analysis of BrCYP83B1 Gene in Chinese Cabbage [J]. Biotechnology Bulletin, 2024, 40(6): 152-160. |
| [2] | YAN Huan-huan, SHANG Yi-tong, WANG Li-hong, TIAN Xue-qin, LIAO Hai-yan, ZENG Bin, HU Zhi-hong. Heterologous Biosynthesis of Cordycepin in Aspergillus oryzae [J]. Biotechnology Bulletin, 2024, 40(6): 290-298. |
| [3] | CHANG Xue-rui, WANG Tian-tian, WANG Jing. Identification and Analysis of E2 Gene Family in Pepper(Capsicum annuum L.) [J]. Biotechnology Bulletin, 2024, 40(6): 238-250. |
| [4] | LIU Rong, TIAN Min-yu, LI Guang-ze, TAN Cheng-fang, RUAN Ying, LIU Chun-lin. Identification and Induced-expression Analysis of REVEILLE Family in Brassica napus L. [J]. Biotechnology Bulletin, 2024, 40(6): 161-171. |
| [5] | WU Ze-hang, YANG Zhong-yi, YAN Yi-cheng, JIA Yong-hong, WU Yue-yan, XIE Xiao-hong. Cloning and Functional Analysis of Flavonoid 3'-hydroxylase(F3'H)Gene in Rhododendron hybridum Hort [J]. Biotechnology Bulletin, 2024, 40(6): 251-259. |
| [6] | LI Jia-xin, LI Hong-yan, LIU Li-e, ZHANG Tian, ZHOU Wu. Identification and Expression Analysis of the NRAMP Family in Seabuckthorn Under Lead Stress [J]. Biotechnology Bulletin, 2024, 40(5): 191-202. |
| [7] | PAN Ping-ping, XU Zhi-hao, ZHANG Yi-wen, LI Qing, WANG Zhong-hua. Prokaryotic Expression, Subcellular Localization and Expression Analysis of PcCHS Gene from Polygonatum cyrtonema Hua [J]. Biotechnology Bulletin, 2024, 40(5): 280-289. |
| [8] | ZHANG Zhen, LI Qing, XU Jing, CHEN Kai-yuan, ZHANG Chun-zhi, ZHU Guang-tao. Construction and Application of Potato Mitochondrial Targeted Expression Vector [J]. Biotechnology Bulletin, 2024, 40(5): 66-73. |
| [9] | LOU Yin, GAO Hao-jun, WANG Xi, NIU Jing-ping, WANG Min, DU Wei-jun, YUE Ai-qin. Identification and Expression Pattern Analysis of GmHMGS Gene in Soybean [J]. Biotechnology Bulletin, 2024, 40(4): 110-121. |
| [10] | CHEN Chun-lin, LI Bai-xue, LI Jin-ling, DU Qing-jie, LI Meng, XIAO Huai-juan. Identification and Expression Analysis of Epidermal Patterning Factor (EPF) Genes in Cucumis melo [J]. Biotechnology Bulletin, 2024, 40(4): 130-138. |
| [11] | XIAO Ya-ru, JIA Ting-ting, LUO Dan, WU Zhe, LI Li-xia. Cloning and Expression Analysis of CsERF025L Transcription Factor in Cucumber [J]. Biotechnology Bulletin, 2024, 40(4): 159-166. |
| [12] | YANG Wei-cheng, SUN Yan, YANG Qian, WANG Zhuang-lin, MA Ju-hua, XUE Jin-ai, LI Run-zhi. Genome-wide Identification of the FAX family in Gossypium hirsutum and Functional Analysis of GhFAX1 [J]. Biotechnology Bulletin, 2024, 40(3): 155-169. |
| [13] | YANG Yan, HU Yang, LIU Ni-ru, YIN Lu, YANG Rui, WANG Peng-fei, MU Xiao-peng, ZHANG Shuai, CHENG Chun-zhen, ZHANG Jian-cheng. Cloning and Functional Analysis of MbbZIP43 Gene in ‘Hongmantang’ Red-flesh Apple [J]. Biotechnology Bulletin, 2024, 40(2): 146-159. |
| [14] | REN Yan-jing, ZHANG Lu-gang, ZHAO Meng-liang, LI Jiang, SHAO Deng-kui. cDNA Yeast Library Construction of Chinese Cabbage Seeds and Screening and Analysis of BrTTG1 Interacting Proteins [J]. Biotechnology Bulletin, 2024, 40(2): 223-232. |
| [15] | ZHU Yi, LIU Tang-jing, GONG Guo-yi, ZHANG Jie, WANG Jin-fang, ZHANG Hai-ying. Cloning and Expression Analysis of ClPP2C3 in Citrullus lanatus [J]. Biotechnology Bulletin, 2024, 40(1): 243-249. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||