








Biotechnology Bulletin ›› 2024, Vol. 40 ›› Issue (6): 281-289.doi: 10.13560/j.cnki.biotech.bull.1985.2024-0121
Previous Articles Next Articles
WANG Qiu-yue1( ), DUAN Peng-liang1, LI Hai-xiao2, LIU Ning1(
), DUAN Peng-liang1, LI Hai-xiao2, LIU Ning1( ), CAO Zhi-yan1,2(
), CAO Zhi-yan1,2( ), DONG Jin-gao1,2
), DONG Jin-gao1,2
Received:2024-01-31
Online:2024-06-26
Published:2024-05-14
Contact:
LIU Ning, CAO Zhi-yan
E-mail:937813463@qq.com;lning121@126.com;caozhiyan@hebau.edu.cn
WANG Qiu-yue, DUAN Peng-liang, LI Hai-xiao, LIU Ning, CAO Zhi-yan, DONG Jin-gao. Construction of cDNA Library of Setosphaeria turcica and Screening of Transcription Factor StMR1 Interacting Proteins[J]. Biotechnology Bulletin, 2024, 40(6): 281-289.
| 引物名称 Primer name | 引物序列 Primer sequence(5'-3') |
|---|---|
| pGBKT7-StMR1-F | TGGCCATGGAGGCCGAATTCCCGGGATGGTTTTCTGCACATACTGC |
| pGBKT7-StMR1-R | GGGGTTATGCTAGTTATGCGGCCGCTTAGCCGCGCGCAAGCTTTT |
| pGADT7-135640--F | CGACGTACCAGATTACGCTCATATGATGGCAGAGAAGAAGCAGCGC |
| pGADT7-135640-R | TGCAGCTCGAGCTCGATGGATCCTTATTGCTTCAGCGCGAGCA |
| pGADT7-169112-F | CGACGTACCAGATTACGCTCATATGATGCACCTCAACGACTTCCC |
| pGADT7-169112-R | TGCAGCTCGAGCTCGATGGATCCTCACGTCTGGATGGAATGGA |
| pGADT7-165582-F | TGGCCATGGAGGCCAGTGAATTCATGACATCACGACTCATCACC |
| pGADT7-165582-R | CTGCAGCTCGAGCTCGATGGATCCTTACCACTTCTCAACCG |
| M 13-F | GTAAAACGACGGCCAG |
| M 13-R | CAGGAAACAGCTATGAC |
| T7 | TAATACGACTCACTATAGGGC |
| 3-AD | AGATGGTGCACGATGCACAG |
Table 1 Primers used in the experiment
| 引物名称 Primer name | 引物序列 Primer sequence(5'-3') |
|---|---|
| pGBKT7-StMR1-F | TGGCCATGGAGGCCGAATTCCCGGGATGGTTTTCTGCACATACTGC |
| pGBKT7-StMR1-R | GGGGTTATGCTAGTTATGCGGCCGCTTAGCCGCGCGCAAGCTTTT |
| pGADT7-135640--F | CGACGTACCAGATTACGCTCATATGATGGCAGAGAAGAAGCAGCGC |
| pGADT7-135640-R | TGCAGCTCGAGCTCGATGGATCCTTATTGCTTCAGCGCGAGCA |
| pGADT7-169112-F | CGACGTACCAGATTACGCTCATATGATGCACCTCAACGACTTCCC |
| pGADT7-169112-R | TGCAGCTCGAGCTCGATGGATCCTCACGTCTGGATGGAATGGA |
| pGADT7-165582-F | TGGCCATGGAGGCCAGTGAATTCATGACATCACGACTCATCACC |
| pGADT7-165582-R | CTGCAGCTCGAGCTCGATGGATCCTTACCACTTCTCAACCG |
| M 13-F | GTAAAACGACGGCCAG |
| M 13-R | CAGGAAACAGCTATGAC |
| T7 | TAATACGACTCACTATAGGGC |
| 3-AD | AGATGGTGCACGATGCACAG |

Fig. 2 Total RNA extraction(A)and electrophoresis results of mRNA(B)and ds cDNA(C) (A) M: DL 5000 marker, 1-9: RNA;(B-C) M: DL 2000 marker, 1: mRNA,2: ds cDNA

Fig. 3 Identification of library capacity and insert recombination rate A: Identification of primary library capacity. B: 1-24 detection of primary library insert. C: Identification of the secondary library capacity of the nuclear system. D: 1-24 detection of nuclear system secondary library insert. M: DL 2000 marker
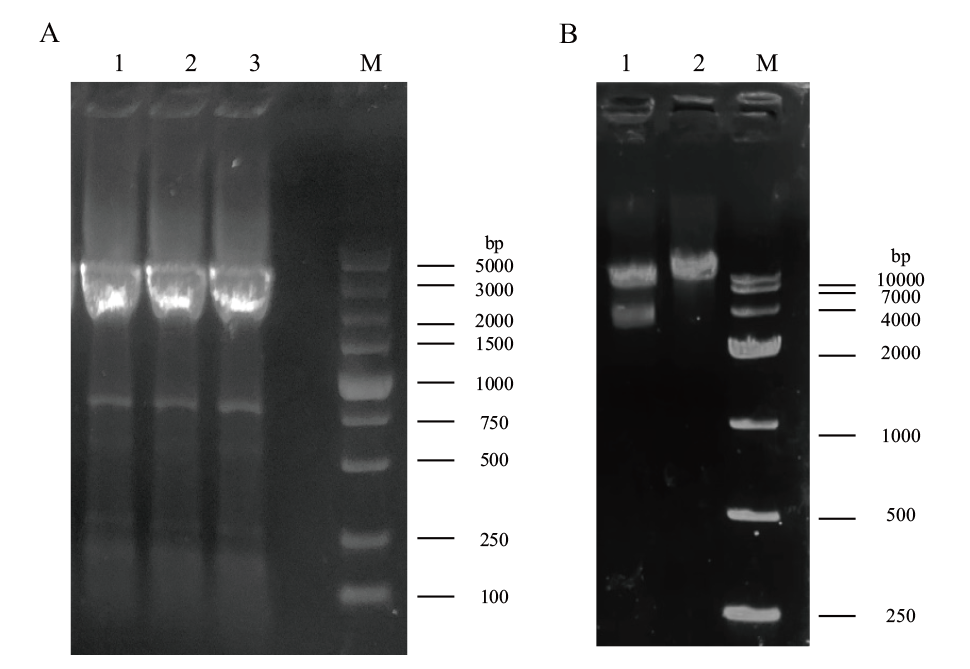
Fig. 4 Construction of bait vector pGBKT7-StMR1 A: Target fragment amplification(1-3: target fragment; M: DL 5000 marker). B: Verification of double digestion of bait vectors(1: digestion; 2: double single digestion; M: DL 10000 marker)
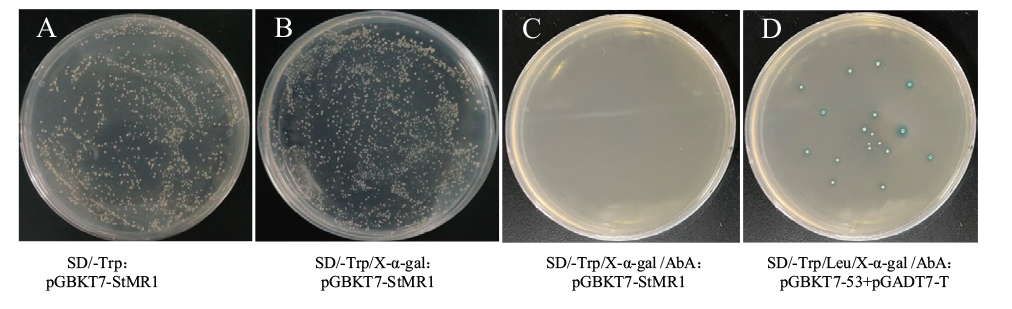
Fig. 5 Detection of toxicity and self-activating activity of bait vector pGBKT7-StMR1 A: SD/-Trp: pGBKT7-StMR1; B: SD/-Trp/X-α-gal: pGBKT7-StMR1; C: SD/-Trp/X-α-gal /AbA: pGBKT7-StMR1; D: SD/-Trp/Leu/X-α-gal/AbA: pGBKT7-53+pGADT7-T
| 基因号 Gene ID | 基因登录号 GenBank No. | 片段大小 Fragment size/bp | 编码蛋白 Encoded protein | 结构域 Pfam |
|---|---|---|---|---|
| SETTUDRAFT-165582 | XM_008032228.1 | 1 396 | 短链脱氢/还原酶 Short chain dehydrogenase/reductase | 短链脱氢酶|烯酰基-(酰基载体蛋白)还原酶|氪域 |
| SETTUDRAFT-135640 | XM_008027394.1 | 1 548 | 糖基转移酶 Glycosyltransferase | 酵母中ALG2是一种1,3-甘露糖基转移酶 |
| SETTUDRAFT-169112 | XM_008027772.1 | 1 920 | 富含亮氨酸重复序列蛋白 Leucine-rich repeat | LRR_AMN1;富含亮氨酸的重复结构 |
Table 2 StMR1 candidate protein
| 基因号 Gene ID | 基因登录号 GenBank No. | 片段大小 Fragment size/bp | 编码蛋白 Encoded protein | 结构域 Pfam |
|---|---|---|---|---|
| SETTUDRAFT-165582 | XM_008032228.1 | 1 396 | 短链脱氢/还原酶 Short chain dehydrogenase/reductase | 短链脱氢酶|烯酰基-(酰基载体蛋白)还原酶|氪域 |
| SETTUDRAFT-135640 | XM_008027394.1 | 1 548 | 糖基转移酶 Glycosyltransferase | 酵母中ALG2是一种1,3-甘露糖基转移酶 |
| SETTUDRAFT-169112 | XM_008027772.1 | 1 920 | 富含亮氨酸重复序列蛋白 Leucine-rich repeat | LRR_AMN1;富含亮氨酸的重复结构 |
| [1] | Mideros SX, Chung CL, Wiesner-Hanks T, et al. Determinants of virulence and in vitro development colocalize on a genetic map of Setosphaeria turcica[J]. Phytopathology, 2018, 108(2): 254-263. |
| [2] | Perkins JM, Pedersen WL. Disease development and yield losses associated with northern leaf blight on corn[J]. Plant Dis, 1987, 71(10): 940. |
| [3] | Dong JG, Fan YS, Gui XM, et al. Geographic distribution and genetic analysis of physiological races of Setosphaeria turcica in northern China[J]. Am J Agric Biol Sci, 2008, 3(1): 389-398. |
| [4] | 曹志艳, 贾慧, 朱显明, 等. DHN黑色素与玉米大斑病菌附着胞膨压形成的关系[J]. 中国农业科学, 2011, 44(5): 925-932. |
| Cao ZY, Jia H, Zhu XM, et al. Relationship between DHN melanin and formation of appressorium turgor pressure of Setosphaeria turcica[J]. Sci Agric Sin, 2011, 44(5): 925-932. | |
| [5] | Guo XY, Liu N, Liu BH, et al. Melanin, DNA replication, and autophagy affect appressorium development in Setosphaeria turcica by regulating glycerol accumulation and metabolism[J]. J integr Agric, 2022, 21(3): 762-773. |
| [6] | Zhang ZX, Jia H, Liu N, et al. The zinc finger protein StMR1 affects the pathoenicity and melanin synthesis of setosphaeria turcica and directly regulates the expression of DHN melanin synthesis pathway genes[J]. Mol Microbiol, 2022, 117(2): 261-273. |
| [7] | Sousa Melo B, Voltan AR, Arruda W, et al. Morphological and molecular aspects of sclerotial development in the phytopathogenic fungus Sclerotinia sclerotiorum[J]. Microbiol Res, 2019, 229: 126326. |
| [8] | Stappers MHT, Clark AE, Aimanianda V, et al. Recognition of DHN-melanin by a C-type lectin receptor is required for immunity to Aspergillus[J]. Nature, 2018, 555(7696): 382-386. |
| [9] |
Tsuji G, Kenmochi Y, Takano Y, et al. Novel fungal transcriptional activators, Cmr1p of Colletotrichum lagenarium and pig1p of Magnaporthe grisea, contain Cys2His2 zinc finger and Zn(II)2Cys6 binuclear cluster DNA-binding motifs and regulate transcription of melanin biosynthesis genes in a developmentally specific manner[J]. Mol Microbiol, 2000, 38(5): 940-954.
pmid: 11123670 |
| [10] | 竺思仪. 稻瘟病菌黑色素合成和调控相关基因的功能研究[D]. 杭州: 浙江大学, 2021. |
| Zhu SY. The study on melanin synthesis and regulation related genes in Magnaporthe oryzae[D]. Hangzhou: Zhejiang University, 2021. | |
| [11] | Langfelder K, Jahn B, Gehringer H, et al. Identification of a polyketide synthase gene(pksP)of Aspergillus fumigatus involved in conidial pigment biosynthesis and virulence[J]. Med Microbiol Immunol, 1998, 187(2): 79-89. |
| [12] | Lin SY, Okuda S, Ikeda K, et al. LAC2 encoding a secreted laccase is involved in appressorial melanization and conidial pigmentation in Colletotrichum orbiculare[J]. Mol Plant Microbe Interact, 2012, 25(12): 1552-1561. |
| [13] |
Eliahu N, Igbaria A, Rose ms, et al. Melanin biosynthesis in the maize pathogen Cochliobolus heterostrophus depends on two mitogen-activated protein kinases, Chk1 and Mps1, and the transcription factor Cmr1[J]. Eukaryot Cell, 2007, 6(3): 421-429.
doi: 10.1128/EC.00264-06 pmid: 17237364 |
| [14] | Kihara J, Moriwaki A, Tanaka N, et al. Characterization of the BMR1 gene encoding a transcription factor for melanin biosynthesis genes in the phytopathogenic fungus Bipolaris oryzae[J]. FEMS Microbiol Lett, 2008, 281(2): 221-227. |
| [15] | Cho Y, Srivastava A, Ohm RA, et al. Transcription factor Amr1 induces melanin biosynthesis and suppresses virulence in Alternaria brassicicola[J]. PLoS Pathog, 2012, 8(10): e1002974. |
| [16] | Zhou YJ, Yang L, Wu MD, et al. A single-nucleotide deletion in the transcription factor gene bcsmr1 causes sclerotial-melanogenesis deficiency in Botrytis cinerea[J]. Front Microbiol, 2017, 8: 2492. |
| [17] | Wang YL, Hu XP, Fang YL, et al. Transcription factor VdCmr 1 is required for pigment production, protection from UV irradiation, and regulates expression of melanin biosynthetic genes in Verticillium dahliae[J]. Microbiology, 2018, 164(4): 685-696. |
| [18] | Carrillo AJ, Schacht P, Cabrera IE, et al. Functional profiling of transcription factor genes in Neurospora crassa[J]. G3, 2017, 7(9): 2945-2956. |
| [19] | 沈竹, 曹勤红. 酵母双杂交及其衍生技术应用研究进展[J]. 农业生物技术学报, 2022, 12: 2425-2433. |
| Shen Z, Cao QH. Research progress on application of yeast two hybrid system and y2h-derivated techniques[J]. J Agric Biotechnol, 2022, 12: 2425-2433. | |
| [20] |
Choi sG, Richardson A, Lambourne L, et al. Protein interactomics by two-hybrid methods[J]. Methods Mol Biol, 2018, 1794: 1-14.
doi: 10.1007/978-1-4939-7871-7_1 pmid: 29855947 |
| [21] | 韦吉, 栾宏瑛, 王荟洁, 等. 致病疫霉诱导的马铃薯酵母双杂交文库构建及无毒蛋白PiAVR3b寄主靶标筛选[J]. 植物保护, 2022, 4: 114-122, 130. |
| Wei J, Luan HY, Wang HJ, et al. Construction of yeast two-hybrid cDNA library induced by Phytophthorainfestans and screening host targets for avirulence protein PiAVR3b in potato[J]. Plant Prot, 2022, 4: 114-122, 130. | |
| [22] | 许艳, 李冉, 宋健, 等. 大丽轮枝菌及激素处理后棉花酵母双杂交文库的构建[J]. 植物保护, 2021, 47(2): 46-55. |
| Xu Y, Li R, Song J, et al. Construction of a sea-island cotton yeast two-hybrid library under the conditions of infection with Verticillium dahliae and treatment with hormones[J]. Plant Prot, 2021, 47(2): 46-55. | |
| [23] | Zhu JK. Salt and drought stress signal transduction in plants[J]. Annu Rev Plant Biol, 2002, 53: 247-273. |
| [24] | 李舒文, 董笛, 李殷睿智, 等. 蒺藜苜蓿酵母杂交cDNA文库的构建与分析[J]. 分子植物育种, 2021, 19(23): 7861-7866. |
| Li SW, Dong D, Li YRZ, et al. Construction and analysis of a yeast cDNA library from Medicago truncatula[J]. Mol Plant Breed, 2021, 19(23): 7861-7866. | |
| [25] |
Luo XM, Xie CJ, Dong JY, et al. Comparative transcriptome analysis reveals regulatory networks and key genes of microsclerotia formation in the cotton vascular wilt pathogen[J]. Fungal Genet Biol, 2019, 126: 25-36.
doi: S1087-1845(18)30217-2 pmid: 30710746 |
| [26] | Zhang L, Dong DH, Wang JF, et al. A zinc finger protein SlSZP1 protects SlSTOP1 from SlRAE1-mediated degradation to modulate aluminum resistance[J]. New Phytol, 2022, 236(1): 165-181. |
| [27] | Yu SH, Sun QG, Wu JX, et al. Genome-wide identificaiton and characterizaiton of short-chain dehydrogenase/reductase(SDR)gene family in Medicago truncatula[J]. Int J Mol Sci, 2021, 22(17): 9498. |
| [28] |
王帅乐, 肖珂雨, 王维东, 等. 苹果短链脱氢酶基因MdSDR响应腐烂病菌侵染的功能研究[J]. 核农学报, 2022, 36(11): 2158-2165.
doi: 10.11869/j.issn.100-8551.2022.11.2158 |
| Wang SL, Xiao KY, Wang WD, et al. Function exploration of apple short-chain dehydrogenases/reductase in response to valsa mali[J]. J Nucl Agric Sci, 2022, 36(11): 2158-2165. | |
| [29] | Huang LJ, Yang WH, Chen JL, et al. Molecular identification and functional characterization of an environmental stress responsive glutaredoxin gene ROXY1 in Quercus glauca[J]. Plant Physiol Biochem, 2024, 207: 108367. |
| [30] | Li J, Wen JQ, Lease KA, et al. BAK1, an Arabidopsis LRR receptor-like protein kinase, interacts with BRI1 and modulates brassinosteroid signaling[J]. Cell, 2002, 110(2): 213-222. |
| [31] | Gharabli H, Della Gala V, Welner DH. The function of UDP-glycosyltransferases in plants and their possible use in crop protection[J]. Biotechnol Adv, 2023, 67: 108182. |
| [32] |
Jo YK, Park NY, Park SJ, et al. O-GlcNAcylation of ATG4B positively regulates autophagy by increasing its hydroxylase activity[J]. Oncotarget, 2016, 7(35): 57186-57196.
doi: 10.18632/oncotarget.11083 pmid: 27527864 |
| [1] | ZHANG Di, JU Rui, LI Li-mei, WANG Yu-qian, CHEN Rui, WANG Xin-yi. Application of Transcription Factor-based Biosensors in Environmental Analysis [J]. Biotechnology Bulletin, 2024, 40(6): 114-125. |
| [2] | HU Ya-dan, WU Guo-qiang, LIU Chen, WEI Ming. Roles of MYB Transcription Factor in Regulating the Responses of Plants to Stress [J]. Biotechnology Bulletin, 2024, 40(6): 5-22. |
| [3] | WANG Jian, YANG Sha, SUN Qing-wen, CHEN Hong-yu, YANG Tao, HUANG Yuan. Genome-wide Identification and Expression Analysis of bHLH Transcription Factor Family in Dendrobium nobile [J]. Biotechnology Bulletin, 2024, 40(6): 203-218. |
| [4] | WANG Di ZHANG Xiao-yu SONG Yu-xin ZHENG Dong-ran TIAN Jing LI Yu-hua WANG Yu WU Hao. Advances in the Molecular Mechanisms of Plant Tissue Culture and Regeneration Regulated by Totipotency-related Transcription Factors [J]. Biotechnology Bulletin, 2024, 40(6): 23-33. |
| [5] | LI Hui, WEN Yu-fang, WANG Yue, JI Chao, SHI Guo-you, LUO Ying, ZHOU Yong, LI Zhi-min, WU Xiao-yu, YANG You-xin, LIU Jian-ping. Expression Characteristics and Functions of CaPIF4 in Capsicum annuum Under Salt Stress [J]. Biotechnology Bulletin, 2024, 40(4): 148-158. |
| [6] | XIAO Ya-ru, JIA Ting-ting, LUO Dan, WU Zhe, LI Li-xia. Cloning and Expression Analysis of CsERF025L Transcription Factor in Cucumber [J]. Biotechnology Bulletin, 2024, 40(4): 159-166. |
| [7] | GUO Chun, SONG Gui-mei, YAN Yan, DI Peng, WANG Ying-ping. Genome Wide Identification and Expression Analysis of the bZIP Gene Family in Panax quinquefolius [J]. Biotechnology Bulletin, 2024, 40(4): 167-178. |
| [8] | XIE Qian, JIANG Lai, HE Jin, LIU Ling-ling, DING Ming-yue, CHEN Qing-xi. Regulatory Genes Mining Related to Transcriptome Sequencing and Phenolic Metabolism Pathway of Canarium album Fruit with Different Fresh Food Quality [J]. Biotechnology Bulletin, 2024, 40(3): 215-228. |
| [9] | CHEN Yan-mei. Crosstalk Between Different Post-translational Modifications and Its Regulatory Mechanisms in Plant Growth and Development [J]. Biotechnology Bulletin, 2024, 40(2): 1-8. |
| [10] | YANG Yan, HU Yang, LIU Ni-ru, YIN Lu, YANG Rui, WANG Peng-fei, MU Xiao-peng, ZHANG Shuai, CHENG Chun-zhen, ZHANG Jian-cheng. Cloning and Functional Analysis of MbbZIP43 Gene in ‘Hongmantang’ Red-flesh Apple [J]. Biotechnology Bulletin, 2024, 40(2): 146-159. |
| [11] | LU Yu-dan, LIU Xiao-chi, FENG Xin, CHEN Gui-xin, CHEN Yi-ting. Identification of the Kiwifruit BBX Gene Family and Analysis of Their Transcriptional Characteristics [J]. Biotechnology Bulletin, 2024, 40(2): 172-182. |
| [12] | XIN Qi, LI Ya-fan, YIN Zheng, ZHANG Xiao-dan, CHEN Ting, LIU Xiao-hua. Identification and Expression Analysis of the CBL-CIPK Gene Family in Sugarcane [J]. Biotechnology Bulletin, 2024, 40(2): 197-211. |
| [13] | REN Yan-jing, ZHANG Lu-gang, ZHAO Meng-liang, LI Jiang, SHAO Deng-kui. cDNA Yeast Library Construction of Chinese Cabbage Seeds and Screening and Analysis of BrTTG1 Interacting Proteins [J]. Biotechnology Bulletin, 2024, 40(2): 223-232. |
| [14] | ZOU Xiu-wei, YUE Jia-ni, LI Zhi-yu, DAI Liang-ying, LI Wei. Functional Analysis of Rice Heat Shock Transcription Factor HsfA2b Regulating the Resistance to Abiotic Stresses [J]. Biotechnology Bulletin, 2024, 40(2): 90-98. |
| [15] | ZHOU Hui-wen, WU Lan-hua, HAN De-peng, ZHENG Wei, YU Pao-lan, WU Yang, XIAO Xiao-jun. Genome-wide Association Study of Seed Glucosinolate Content in Brassica napus [J]. Biotechnology Bulletin, 2024, 40(1): 222-230. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||