








Biotechnology Bulletin ›› 2024, Vol. 40 ›› Issue (6): 330-342.doi: 10.13560/j.cnki.biotech.bull.1985.2023-1005
Previous Articles Next Articles
QIN Jian1,2( ), LI Zhen-yue1, HE Lang1, LI Jun-ling1, ZHANG Hao1, DU Rong1(
), LI Zhen-yue1, HE Lang1, LI Jun-ling1, ZHANG Hao1, DU Rong1( )
)
Received:2023-10-30
Online:2024-06-26
Published:2024-06-24
Contact:
DU Rong
E-mail:qinjian969@163.com;drdurong@163.com
QIN Jian, LI Zhen-yue, HE Lang, LI Jun-ling, ZHANG Hao, DU Rong. Change of Single-cell Transcription Profile and Analysis of Intercellular Communication in Myogenic Cell Differentiation[J]. Biotechnology Bulletin, 2024, 40(6): 330-342.
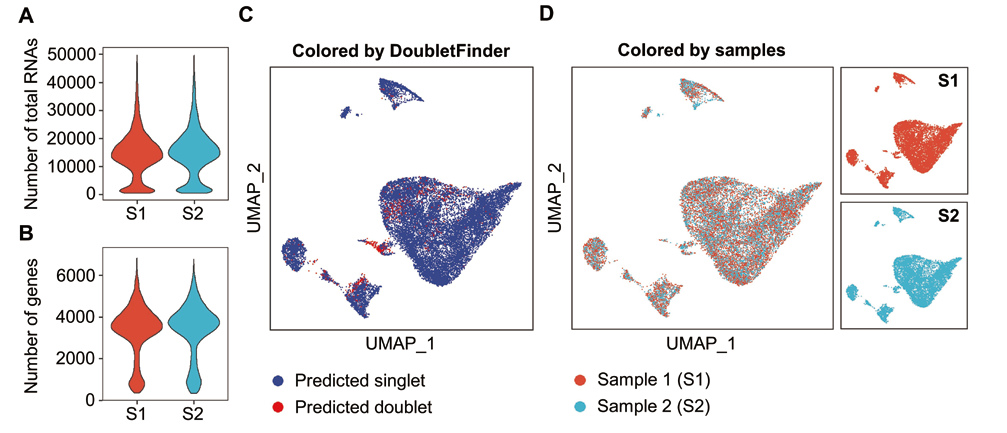
Fig. 1 Quality control and sample integration analysis of scRNA-seq data from bovine myogenic cell samples A: A violin plot shows the number of total RNAs in the remaining high-quality cells of two samples after quality control and filtration. B: A violin plot shows the number of total genes in the remaining high-quality cells of two samples after quality control and filtration. C: A UMAP dimension reduction graph shows the singlet or doublet(single or double cells)property prediction results of two samples after integrated analysis(Blue indicates singlet and red indicates doublet). D: UMAP dimension reduction graphs show the singlet presentations of two samples after filtering the doublet(The left shows the combined display and the right shows the separate display. Red indicates sample 1 and blue indicates sample 2)

Fig. 2 Cell type profile of bovine myogenic cell samples delineated by scRNA-seq analysis A: A UMAP dimension reduction graph shows the 12 cell clusters obtained from scRNA-seq analysis(Different colors indicate the different cell clusters). B: A dot plot shows the expression levels of marker genes in each cell cluster(Color scale indicates the average expression level of a certain gene in the cell clusters, and dot size indicates the percentage of cells expressing a certain gene). C: A heatmap shows the correlations among 12 cell clusters based on the differential gene expression levels(Color scale indicates correlation coefficient). D: A UMAP dimension reduction graph shows the 12 cell clusters are annotated into 4 cell types based on the marker gene and correlation analysis(Different colors indicate the different cell types). E: UMAP graphs combined with FeaturePlots show the expression levels of marker genes for each cell type(Color scale indicates the gene expression level)

Fig. 3 Cell subclusters and differential gene expression profile comparison of bovine Myoblasts A: Circle graphs show the proportions of four cell types from two samples of bovine myogenic cells. B: A UMAP dimension reduction graph shows the Myoblasts used to divide subclusters after the two samples are combined(Blue indicates Myoblasts). C: A UMAP dimension reduction graph shows the six Myoblasts subclusters(Different colors indicate the different cell subclusters). D: A dot plot shows the comparison of the representative differential gene expression profiles of the Myoblasts subclusters(Color scale indicates the average expression level of a certain gene in the cell subclusters, and dot size indicates the percentage of cells expressing a certain gene)
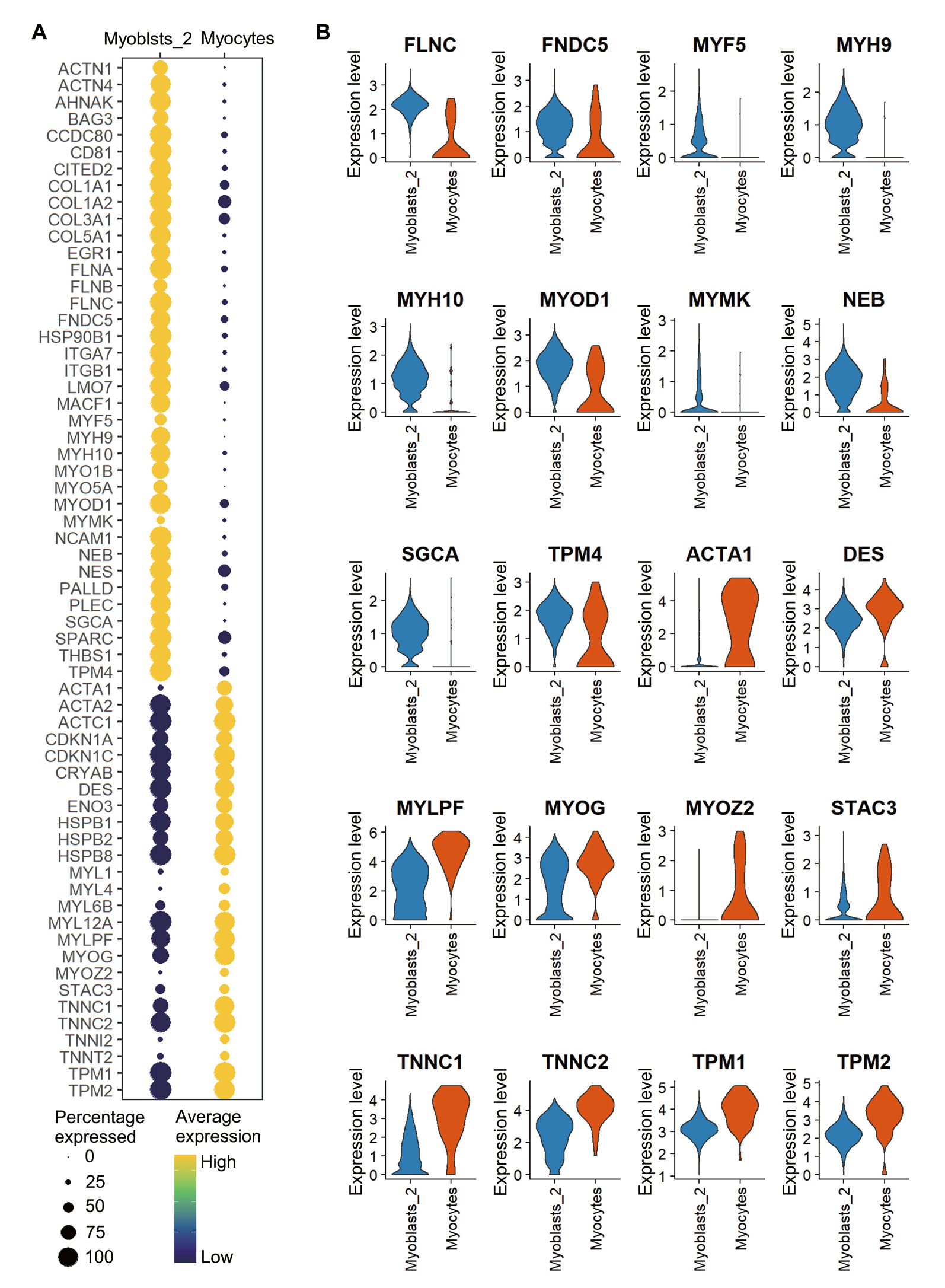
Fig. 4 Comparison of the major muscle-related differential genes between Myoblasts_2 and Myocytes A: A dot plot shows the expression profile of the major muscle-related differential genes between Myoblasts_2 and Myocytes(Color scale indicates the average expression level of a certain gene in the cell cluster or subcluster, and dot size indicates the percentage of cells expressing a certain gene). B: Violin plots show the expression levels and cell distributions of some muscle-specific differential genes in Myoblasts_2 and Myocytes(Horizontal coordinate indicates the cell type and vertical coordinate indicates the gene expression level. The width of the violin indicates the cell density with the corresponding expression level, the larger the width, the more cells at the expression level)

Fig. 5 GO enrichment and PPI analysis of the Myoblasts_2 and Myocytes differential genes in muscle-related biological processes A: Muscle-related GO terms enriched by the up-regulated genes in Myoblasts_2. B: Muscle-related GO terms enriched by the up-regulated genes in Myocytes. C: PPI analysis of genes in muscle-related GO terms enriched by Myoblasts_2 and Myocytes(Yellow and green indicate the up-regulated genes respectively in Myoblasts_2 and Myocytes)

Fig. 6 Analyses of ligand-receptor for intercellular communication of various cell types in bovine myogenic cell samples A: A UMAP dimension reduction graph shows the position relationship of the six myoblasts subclusters projected on the cell clusters(Different colors indicate the different cell clusters or subclusters). B: A circle plot shows the interaction strength between various cell types(The size of the surrounding circular nodes indicates the cell number in a certain cell type. The color indicates which cell is the sender, the cell that sends the arrow is the sender, and the cell that the arrow points to is the receiver. The thickness of the line indicates the interaction strength). C: A dot plot shows the major ligand-receptor pairs involved in intercellular communication(Dot size indicates the P-Value, and dot color indicates the ligand-receptor communication probability). D: A violin plot shows the expression levels and cell distributions of PTN ligand and SDC/NCL receptor genes in each cell type(Horizontal coordinate indicates the cell type and vertical coordinate indicates the gene expression level. The width of the violin indicates the cell density with the corresponding expression level, the larger the width, the more cells at the expression level)
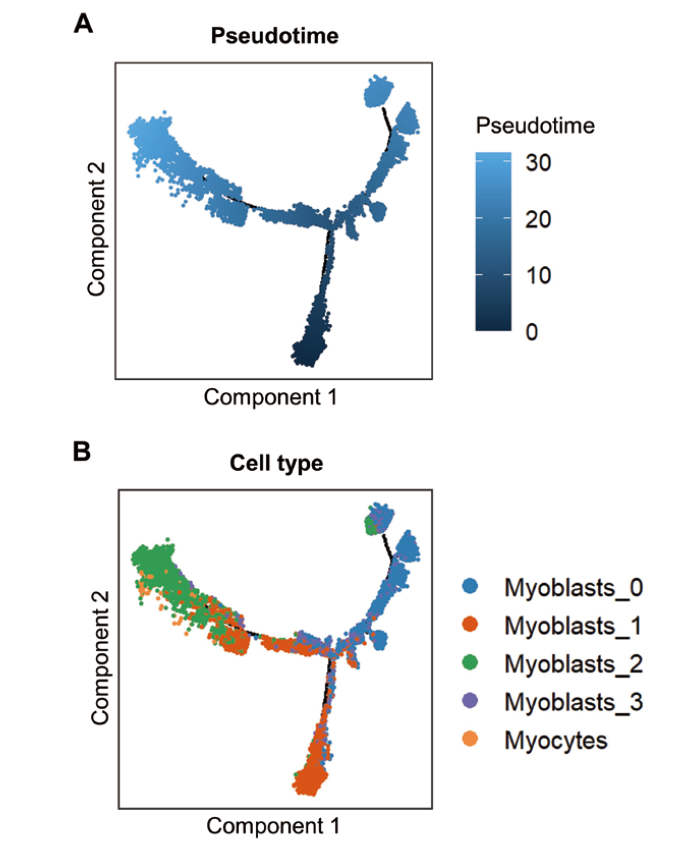
Fig. 7 Differentiation trajectory of bovine myogenic cell subclusters based on pseudotime analysis A: Cell trajectory plot displayed by pseudotime(The color from dark to light indicates the beginning to end of pseudotime). B: Cell trajectory plot displayed by cell type(Different colors indicate different cell types)
| [1] | Li JJ, Pei YL, Zhou R, et al. Regulation of RNA N6-methyladenosine modification and its emerging roles in skeletal muscle development[J]. Int J Biol Sci, 2021, 17(7): 1682-1692. |
| [2] | Rodríguez-Fdez S, Bustelo XR. Rho GTPases in skeletal muscle development and homeostasis[J]. Cells, 2021, 10(11): 2984. |
| [3] |
Weskamp K, Olwin BB, Parker R. Post-transcriptional regulation in skeletal muscle development, repair, and disease[J]. Trends Mol Med, 2021, 27(5): 469-481.
doi: 10.1016/j.molmed.2020.12.002 pmid: 33384234 |
| [4] | Cao JY, Spielmann M, Qiu XJ, et al. The single-cell transcriptional landscape of mammalian organogenesis[J]. Nature, 2019, 566(7745): 496-502. |
| [5] | Cai CC, Yue Y, Yue BL. Single-cell RNA sequencing in skeletal muscle developmental biology[J]. Biomed Pharmacother, 2023, 162: 114631. |
| [6] |
Tang FC, Barbacioru C, Wang YZ, et al. mRNA-Seq whole-transcriptome analysis of a single cell[J]. Nat Methods, 2009, 6(5): 377-382.
doi: 10.1038/nmeth.1315 pmid: 19349980 |
| [7] |
Zhang XN, Li TQ, Liu F, et al. Comparative analysis of droplet-based ultra-high-throughput single-cell RNA-seq systems[J]. Mol Cell, 2019, 73(1): 130-142.e5.
doi: S1097-2765(18)30880-3 pmid: 30472192 |
| [8] |
Xi HB, Langerman J, Sabri S, et al. A human skeletal muscle atlas identifies the trajectories of stem and progenitor cells across development and from human pluripotent stem cells[J]. Cell Stem Cell, 2020, 27(1): 158-176.e10.
doi: S1934-5909(20)30156-9 pmid: 32396864 |
| [9] | Cai SF, Hu B, Wang XY, et al. Integrative single-cell RNA-seq and ATAC-seq analysis of myogenic differentiation in pig[J]. BMC Biol, 2023, 21(1): 19. |
| [10] | McKellar DW, Walter LD, Song LT, et al. Large-scale integration of single-cell transcriptomic data captures transitional progenitor states in mouse skeletal muscle regeneration[J]. Commun Biol, 2021, 4(1): 1280. |
| [11] | Lyu PC, Qi YM, Tu ZJ, et al. Single-cell RNA sequencing reveals heterogeneity of cultured bovine satellite cells[J]. Front Genet, 2021, 12: 742077. |
| [12] | Cai CC, Wan P, Wang H, et al. Transcriptional and open chromatin analysis of bovine skeletal muscle development by single-cell sequencing[J]. Cell Prolif, 2023, 56(9): e13430. |
| [13] |
Wang LS, Gao PD, Li CY, et al. A single-cell atlas of bovine skeletal muscle reveals mechanisms regulating intramuscular adipogenesis and fibrogenesis[J]. J Cachexia Sarcopenia Muscle, 2023, 14(5): 2152-2167.
doi: 10.1002/jcsm.13292 pmid: 37439037 |
| [14] |
Satija R, Farrell JA, Gennert D, et al. Spatial reconstruction of single-cell gene expression data[J]. Nat Biotechnol, 2015, 33(5): 495-502.
doi: 10.1038/nbt.3192 pmid: 25867923 |
| [15] |
Butler A, Hoffman P, Smibert P, et al. Integrating single-cell transcriptomic data across different conditions, technologies, and species[J]. Nat Biotechnol, 2018, 36(5): 411-420.
doi: 10.1038/nbt.4096 pmid: 29608179 |
| [16] |
Stuart T, Butler A, Hoffman P, et al. Comprehensive integration of single-cell data[J]. Cell, 2019, 177(7): 1888-1902.e21.
doi: S0092-8674(19)30559-8 pmid: 31178118 |
| [17] |
McGinnis CS, Murrow LM, Gartner ZJ. DoubletFinder: doublet detection in single-cell RNA sequencing data using artificial nearest neighbors[J]. Cell Syst, 2019, 8(4): 329-337.e4.
doi: S2405-4712(19)30073-0 pmid: 30954475 |
| [18] |
Yu GC, Wang LG, Han YY, et al. clusterProfiler: an R package for comparing biological themes among gene clusters[J]. OMICS, 2012, 16(5): 284-287.
doi: 10.1089/omi.2011.0118 pmid: 22455463 |
| [19] | Jin SQ, Guerrero-Juarez CF, Zhang LH, et al. Inference and analysis of cell-cell communication using CellChat[J]. Nat Commun, 2021, 12(1): 1088. |
| [20] |
Qiu XJ, Mao Q, Tang Y, et al. Reversed graph embedding resolves complex single-cell trajectories[J]. Nat Methods, 2017, 14(10): 979-982.
doi: 10.1038/nmeth.4402 pmid: 28825705 |
| [21] | Zhang XX, Lan YJ, Xu JY, et al. CellMarker: a manually curated resource of cell markers in human and mouse[J]. Nucleic Acids Res, 2019, 47(D1): D721-D728. |
| [22] | Hao DD, Becker N, Mückter E, et al. In vitro model of human skeletal muscle tissue for the study of resident macrophages and stem cells[J]. Biology, 2022, 11(6): 936. |
| [23] |
Chong JX, Talbot JC, Teets EM, et al. Mutations in MYLPF cause a novel segmental amyoplasia that manifests as distal arthrogryposis[J]. Am J Hum Genet, 2020, 107(2): 293-310.
doi: S0002-9297(20)30202-0 pmid: 32707087 |
| [24] |
Hall MN, Griffin CA, Simionescu A, et al. Distinct roles for classical nuclear import receptors in the growth of multinucleated muscle cells[J]. Dev Biol, 2011, 357(1): 248-258.
doi: 10.1016/j.ydbio.2011.06.032 pmid: 21741962 |
| [25] |
Mancini M, Magnani E, Macchi F, et al. The multi-functionality of UHRF1: epigenome maintenance and preservation of genome integrity[J]. Nucleic Acids Res, 2021, 49(11): 6053-6068.
doi: 10.1093/nar/gkab293 pmid: 33939809 |
| [26] | Xiao FY, Jiang ZP, Yuan F, et al. Down-regulating NQO1 promotes cellular proliferation in K562 cells via elevating DNA synthesis[J]. Life Sci, 2020, 248: 117467. |
| [27] | Chen J, Chen LD, Hua J, et al. Long-term dynamic compression enhancement TGF-β3-induced chondrogenesis in bovine stem cells: a gene expression analysis[J]. BMC Genom Data, 2021, 22(1): 13. |
| [28] |
Wang Y, Chen L, Ju LG, et al. Novel biomarkers associated with progression and prognosis of bladder cancer identified by co-expression analysis[J]. Front Oncol, 2019, 9: 1030.
doi: 10.3389/fonc.2019.01030 pmid: 31681575 |
| [29] |
Calvo F, Ege N, Grande-Garcia A, et al. Mechanotransduction and YAP-dependent matrix remodelling is required for the generation and maintenance of cancer-associated fibroblasts[J]. Nat Cell Biol, 2013, 15(6): 637-646.
doi: 10.1038/ncb2756 pmid: 23708000 |
| [30] |
Fowler PC, Byrne DJ, O’Sullivan NC. Rare disease models provide insight into inherited forms of neurodegeneration[J]. J Rare Dis Res Treat, 2016, 1(3): 17-21.
pmid: 28603788 |
| [31] | Barbhuiya TK, Fisher M, Boittier ED, et al. Structural investigation of CDCA3-Cdh1 protein-protein interactions using in vitro studies and molecular dynamics simulation[J]. Protein Sci, 2023, 32(3): e4572. |
| [32] |
Fei F, Qu J, Li CY, et al. Role of metastasis-induced protein S100A4 in human non-tumor pathophysiologies[J]. Cell Biosci, 2017, 7: 64.
doi: 10.1186/s13578-017-0191-1 pmid: 29204268 |
| [33] |
Nigdelioglu R, Hamanaka RB, Meliton AY, et al. Transforming growth factor(TGF)-β promotes de novo serine synthesis for collagen production[J]. J Biol Chem, 2016, 291(53): 27239-27251.
doi: 10.1074/jbc.M116.756247 pmid: 27836973 |
| [34] | Cui C, Yin HD, Han SS, et al. Quantitative proteomic and phosphoproteomic analysis of chicken skeletal muscle during embryonic development[J]. Anim Biotechnol, 2023, 34(2): 122-133. |
| [35] | Daudon M, Ramé C, Estienne A, et al. Impact of fibronectin type III domain-containing family in the changes in metabolic and hormonal profiles during peripartum period in dairy cows[J]. Front Vet Sci, 2022, 9: 960778. |
| [36] | Sellathurai J, Nielsen J, Hejbøl EK, et al. Low oxygen tension enhances expression of myogenic genes when human myoblasts are activated from G0 arrest[J]. PLoS One, 2016, 11(7): e0158860. |
| [37] | Noureddine M, Gehmlich K. Structural and signaling proteins in the Z-disk and their role in cardiomyopathies[J]. Front Physiol, 2023, 14: 1143858. |
| [38] | Liu LY, Chen CL, Liu P, et al. MYH10 combines with MYH9 to recruit USP45 by deubiquitinating snail and promotes serous ovarian cancer carcinogenesis, progression, and cisplatin resistance[J]. Adv Sci, 2023, 10(14): e2203423. |
| [39] | Dube DK, Dube S, Abbott L, et al. Identification, characterization, and expression of sarcomeric tropomyosin isoforms in zebrafish[J]. Cytoskeleton, 2017, 74(3): 125-142. |
| [40] | Maggio S, Canonico B, Ceccaroli P, et al. Modulation of the circulating extracellular vesicles in response to different exercise regimens and study of their inflammatory effects[J]. Int J Mol Sci, 2023, 24(3): 3039. |
| [41] |
Paradžik M, Humphries JD, Stojanović N, et al. KANK2 links αVβ5 focal adhesions to microtubules and regulates sensitivity to microtubule poisons and cell migration[J]. Front Cell Dev Biol, 2020, 8: 125.
doi: 10.3389/fcell.2020.00125 pmid: 32195252 |
| [42] |
Lohmeier-Vogel EM, Heeley DH. Biochemical comparison of Tpm1.1(α)and Tpm2.2(β)tropomyosins from rabbit skeletal muscle[J]. Biochemistry, 2016, 55(9): 1418-1427.
doi: 10.1021/acs.biochem.5b01140 pmid: 26863527 |
| [43] |
Wang X. Pleiotrophin: activity and mechanism[J]. Adv Clin Chem, 2020, 98: 51-89.
doi: S0065-2423(20)30015-9 pmid: 32564788 |
| [44] | Poimenidi E, Theodoropoulou C, Koutsioumpa M, et al. Vascular endothelial growth factor A(VEGF-A)decreases expression and secretion of pleiotrophin in a VEGF receptor-independent manner[J]. Vascul Pharmacol, 2016, 80: 11-19. |
| [45] |
Erikson DW, Burghardt RC, Bayless KJ, et al. Secreted phosphoprotein 1(SPP1, osteopontin)binds to integrin alpha v beta 6 on porcine trophectoderm cells and integrin alpha v beta 3 on uterine luminal epithelial cells, and promotes trophectoderm cell adhesion and migration[J]. Biol Reprod, 2009, 81(5): 814-825.
doi: 10.1095/biolreprod.109.078600 pmid: 19571258 |
| [46] | Frank JW, Seo H, Burghardt RC, et al. ITGAV (alpha v integrins)bind SPP1(osteopontin)to support trophoblast cell adhesion[J]. Reproduction, 2017, 153(5): 695-706. |
| [47] | Torrente Y, Bella P, Tripodi L, et al. Role of insulin-like growth factor receptor 2 across muscle homeostasis: implications for treating muscular dystrophy[J]. Cells, 2020, 9(2): 441. |
| [48] | Chen WT, You WJ, Valencak TG, et al. Bidirectional roles of skeletal muscle fibro-adipogenic progenitors in homeostasis and disease[J]. Ageing Res Rev, 2022, 80: 101682. |
| [49] | Hanley CJ, Waise S, Ellis MJ, et al. Single-cell analysis reveals prognostic fibroblast subpopulations linked to molecular and immunological subtypes of lung cancer[J]. Nat Commun, 2023, 14(1): 387. |
| [50] | Nawaz A, Bilal M, Fujisaka S, et al. Depletion of CD206+ M2-like macrophages induces fibro-adipogenic progenitors activation and muscle regeneration[J]. Nat Commun, 2022, 13(1): 7058. |
| [51] | Riparini G, Simone JM, Sartorelli V. FACS-isolation and culture of fibro-adipogenic progenitors and muscle stem cells from unperturbed and injured mouse skeletal muscle[J]. J Vis Exp, 2022(184). DOI: 10.3791/6398 |
| [52] | Cameron A, Wakelin G, Gaulton N, et al. Identification of underexplored mesenchymal and vascular-related cell populations in human skeletal muscle[J]. Am J Physiol Cell Physiol, 2022, 323(6): C1586-C1600. |
| [53] | Kadomatsu T, Endo M, Miyata K, et al. Diverse roles of ANGPTL2 in physiology and pathophysiology[J]. Trends Endocrinol Metab, 2014, 25(5): 245-254. |
| [1] | WU Hao, LIU Zi-wei, ZHENG Ying, DAI Ya-wen, SHI Quan. Study on the Heterogeneity of Human Gingival Mesenchymal Stem Cells at Single Cell Level [J]. Biotechnology Bulletin, 2023, 39(7): 325-332. |
| [2] | KOU Jia-yi, WANG Yu-ling, ZENG Rui-lin, LAN Dao-liang. Application of Single-cell Transcriptome Sequencing in Mammalian [J]. Biotechnology Bulletin, 2022, 38(11): 41-48. |
| [3] | YANG Hong-liang, YUAN Zhen, QIAN Xu-jia-zhi, XU Da-wei. Expression Profile Analysis of Thionin-like Gene Family in Barley [J]. Biotechnology Bulletin, 2022, 38(10): 140-147. |
| [4] | ZHU Qing-yuan, LI Tian-qing. Applications of Single-cell RNA Sequencing in Heart Development,Disease and Medicine [J]. Biotechnology Bulletin, 2021, 37(1): 145-154. |
| [5] | CáO Yán-ting, LIU Yán-feng, LI Jiáng-huá, LIU Long, DU Guo-cheng. ádvánces of Improving the Efficiency of Chemicál Biosynthesis Básed on Cell Subpopulátion Regulátion [J]. Biotechnology Bulletin, 2020, 36(4): 19-25. |
| [6] | WANG Dan-rui, SHEN Wen-li, WEI Zi-yan, WANG Shang, DENG Ye. Applications of Single-cell Sequencing Technology in Microbial Ecology [J]. Biotechnology Bulletin, 2020, 36(10): 237-246. |
| [7] | WANG Na, GONG Na, LIU Guo-li, MA Xiao-ying, YANG Zhen, YANG Tao. Expression Profile Analysis of Maize Resistance Under Osmotic Stress Induced by Secondary Metabolites of Endophytes [J]. Biotechnology Bulletin, 2016, 32(7): 87-92. |
| [8] | PENG Yong, CHEN Shang-wu, MA Hui-qin. Differential Expression Analysis of Gene Expression Profiles During Fruit Development and Ripening of Lycium ruthenicum [J]. Biotechnology Bulletin, 2016, 32(11): 144-151. |
| [9] | Liu Jintao, Wang Xingyi, Fan Li, Deng Xiancun, Liu Xuping, Tan Wensong. Effect of pH Heterogeneity in Large-scale Bioreactor on Fed-batch Culture Process of CHO cells [J]. Biotechnology Bulletin, 2015, 31(10): 236-241. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||