








LIU Xiu-ping1( ), ZHU Xing-yu1(
), ZHU Xing-yu1( ), WANG Guang-yi1,2(
), WANG Guang-yi1,2( )
)
Received:2025-03-17
Online:2025-07-17
Published:2025-07-17
Contact:
WANG Guang-yi
E-mail:liuxp880@tju.edu.cn;gywang@tju.edu.cn
LIU Xiu-ping, ZHU Xing-yu, WANG Guang-yi. Research Progress in Metabolic Engineering for Modification and Syngas-directed Conversion by Acetogenic Clostridium[J]. Biotechnology Bulletin, doi: 10.13560/j.cnki.biotech.bull.1985.2025-0283.

Fig. 1 Wood-Ljungdahl pathwayFdh: Formate dehydrogenase; Fhs: formyl-THF synthetase; FTC: formyl-THF cyclohydrolase; MTDH: methylene-THF dehydrogenase; MetF: methylene-THF reductase; MetR: methyltransferase; CODH/ACS: carbon monoxide dehydrogenase/acetyl-CoA synthase; Ald: acetaldehyde dehydrogenase; AdhE: acetaldehyde/alcohol dehydrogenase; Pta: phosphotransacetylase; Ack: acetate kinase; Aor: aldehyde: ferredoxin oxidoreductase
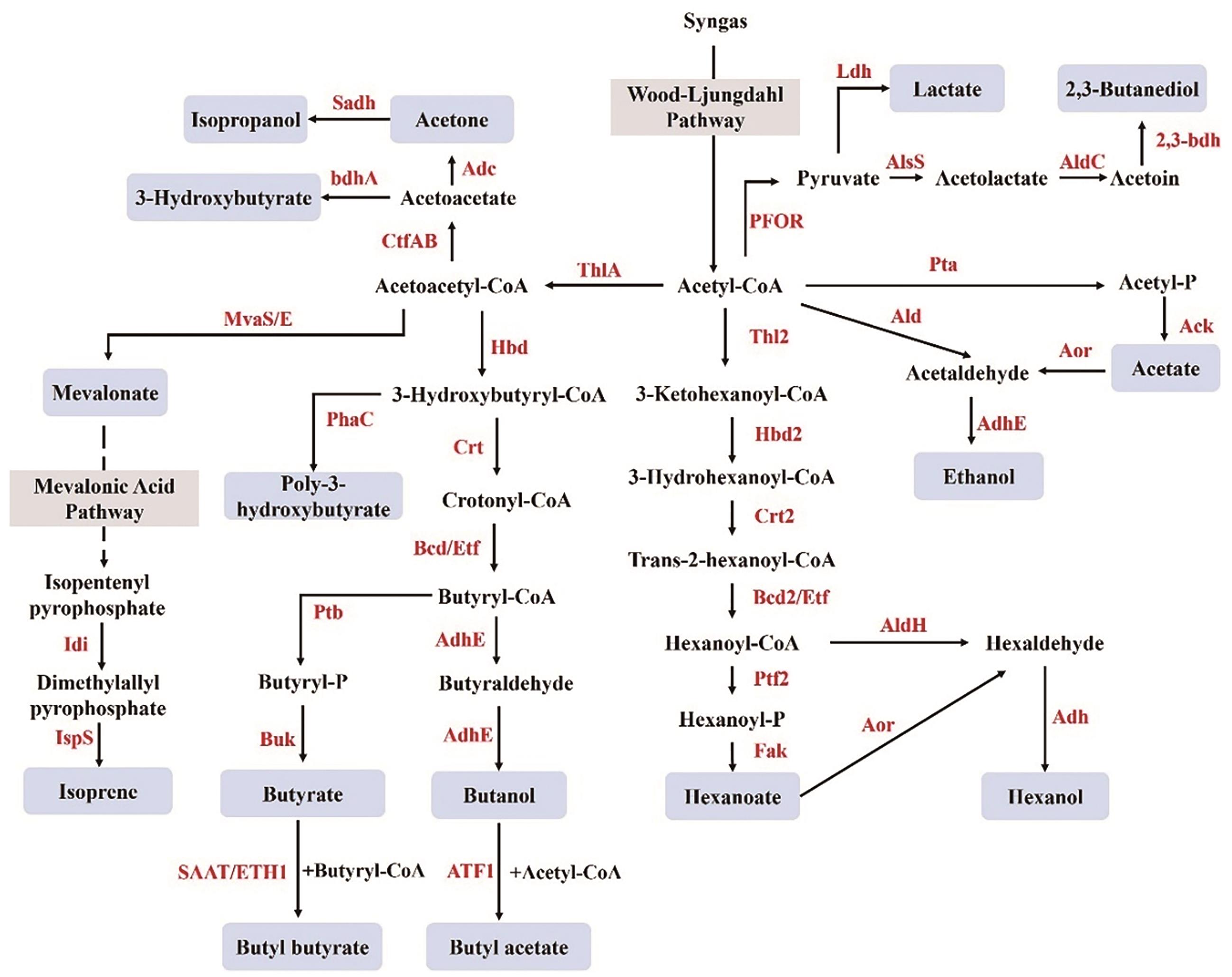
Fig. 3 Metabolic pathways of synthetic products in Acetogenic ClostridiumThlA/Thl2: Thiolase; Hbd/Hbd2: 3-hydroxybutyryl-CoA dehydrogenase; Crt/Crt2: crotonase; Bcd/Bcd2: butyryl-CoA dehydrogenase; AdhE: acetaldehyde/alcohol dehydrogenase; ATF1: alcohol Acetyltransferase; Ptb/Ptf2: Phosphotransferase; Buk: butyrate kinase; SAAT: strawberry Alcohol Acyltransferase; ETH1: ethanol hexanoyltransferase I; MvaS/E: hydroxymethylglutaryl-CoA synthase/reductase; Idi: isopentenyl pyrophosphate isomerase; IspS: isoprene synthase; CtfAB: acetoacetyl-CoA:acetate/butyrate-CoA transferase; Adc: acetoacetate decarboxylase; Sadh: secondary alcohol dehydrogenase; bdhA: 3-hydroxybutyrate dehydrogenase; Pta: Phosphotransacetylase; Ack: Acetate kinase; Ald: Acetaldehyde dehydrogenase; Aor: aldehyde: ferredoxin oxidoreductase; Etf: electron transfer flavoprotein; Fak: fatty acid kinase; AldH: aldehyde dehydrogenasel; Adh: alcohol dehydrogenase; PFOR: pyruvate: ferredoxin oxidoreductase; AlsS: acetolactate synthase; AldC: acetoin decarboxylase; 2,3-Bdh: 2,3-butanediol dehydrogenase; Ldh: lactate dehydrogenase; PhaC: Polyhydroxyalkanoate Synthase
| [1] | Kuang CY. Analysis of Green House Gases and Positive Impact of Replacing Traditional Energy with Clean Energy [C]. E3S Web of Conferences, 2021,241: 2005. |
| [2] | Salehizadeh H, Yan N, Farnood R. Recent advances in microbial CO2 fixation and conversion to value-added products [J]. Chem Eng J, 2020, 390: 124584. |
| [3] | Nguyen T, Hilliard M, Rochelle GT. Amine volatility in CO2 capture [J]. Int J Greenh Gas Control, 2010, 4(5): 707-715. |
| [4] | Sevilla M, Fuertes AB. CO2 adsorption by activated templated carbons [J]. J Colloid Interface Sci, 2012, 366(1): 147-154. |
| [5] | Mustafa J, Farhan M. CO2 separation from flue gases using different types of membranes [J]. J Membra Sci Technol, 2016, 6(2): 1-7. |
| [6] | Xu G, Li L, Yang YP, et al. A novel CO2 cryogenic liquefaction and separation system [J]. Energy, 2012, 42(1): 522-529. |
| [7] | Dashti H, Zhehao Yew L, Lou X. Recent advances in gas hydrate-based CO2 capture [J]. J Nat Gas Sci Eng, 2015, 23: 195-207. |
| [8] | 李婉麒, 杨凤娟, 贾德臣, 等. 合成气的生物利用与定向转化 [J]. 化工进展, 2023. 42(1): 73-85. |
| Li WQ, Yang FJ, Jia DC, et al. Biological utilization and conversion of syngas [J]. Chemical Industry and Engineering Progress, 2023. 42(1): 73-85. | |
| [9] | Köpke M, Held C, Hujer S, et al. Clostridium ljungdahlii represents a microbial production platform based on syngas [J]. Proc Natl Acad Sci USA, 2010, 107(29): 13087-13092. |
| [10] | Patakova P, Kolek J, Sedlar K, et al. Comparative analysis of high butanol tolerance and production in clostridia [J]. Biotechnol Adv, 2018, 36(3): 721-738. |
| [11] | 贾德臣, 姜卫红, 顾阳. 食气梭菌的研究进展 [J]. 微生物学通报, 2019, 46(2): 374-387. |
| Jia DC, Jiang WH, Gu Y. Research progresses in gas-fermenting clostridia [J]. Microbiol China, 2019, 46(2): 374-387. | |
| [12] | Liew F, Martin ME, Tappel RC, et al. Gas fermentation-a flexible platform for commercial scale production of low-carbon-fuels and chemicals from waste and renewable feedstocks [J]. Front Microbiol, 2016, 7: 694. |
| [13] | Grahame DA. Acetate C-C bond formation and decomposition in the anaerobic world: the structure of a central enzyme and its key active-site metal cluster [J]. Trends Biochem Sci, 2003, 28(5): 221-224. |
| [14] | Lee S, Song Y, Choe D, et al. Reconstruction of acetogenesis pathway using short-read sequencing of Clostridium aceticum genome [J]. J Nanosci Nanotechnol, 2015, 15(5): 3852-3861. |
| [15] | Latif H, Zeidan AA, Nielsen AT, et al. Trash to treasure: production of biofuels and commodity chemicals via syngas fermenting microorganisms [J]. Curr Opin Biotechnol, 2014, 27: 79-87. |
| [16] | Zhu HF, Liu ZY, Zhou X, et al. Energy conservation and carbon flux distribution during fermentation of CO or H2/CO2 by Clostridium ljungdahlii [J]. Front Microbiol, 2020, 11: 416. |
| [17] | Müller V. Energy conservation in acetogenic bacteria [J]. Appl Environ Microbiol, 2003, 69(11): 6345-6353. |
| [18] | Müller V, Imkamp F, Biegel E, et al. Discovery of a ferredoxin: NAD+-oxidoreductase (rnf) in Acetobacterium woodii: a novel potential coupling site in acetogens [J]. Ann N Y Acad Sci, 2008, 1125: 137-146. |
| [19] | Tremblay PL, Zhang T, Dar SA, et al. The Rnf complex of Clostridium ljungdahlii is a proton-translocating ferredoxin: NAD+ oxidoreductase essential for autotrophic growth [J]. mBio, 2012, 4(1): e00406-12. |
| [20] | Peters JW, Miller AF, Jones AK, et al. Electron bifurcation [J]. Curr Opin Chem Biol, 2016, 31: 146-152. |
| [21] | Buckel W, Thauer RK. Energy conservation via electron bifurcating ferredoxin reduction and proton/Na+ translocating ferredoxin oxidation [J]. Biochim Biophys Acta BBA Bioenerg, 2013, 1827(2): 94-113. |
| [22] | Wang SN, Huang HY, Moll J, et al. NADP+ reduction with reduced ferredoxin and NADP+ reduction with NADH are coupled via an electron-bifurcating enzyme complex in Clostridium kluyveri [J]. J Bacteriol, 2010, 192(19): 5115-5123. |
| [23] | Wang SN, Huang HY, Kahnt J, et al. NADP-specific electron-bifurcating [FeFe]-hydrogenase in a functional complex with formate dehydrogenase in Clostridium autoethanogenum grown on CO [J]. J Bacteriol, 2013, 195(19): 4373-4386. |
| [24] | Mock J, Zheng YN, Mueller AP, et al. Energy conservation associated with ethanol formation from H2 and CO2 in Clostridium autoethanogenum involving electron bifurcation [J]. J Bacteriol, 2015, 197(18): 2965-2980. |
| [25] | Richter H, Molitor B, Wei H, et al. Ethanol production in syngas-fermenting Clostridium ljungdahlii is controlled by thermodynamics rather than by enzyme expression [J]. Energy Environ Sci, 2016, 9(7): 2392-2399. |
| [26] | Bertsch J, Müller V. Bioenergetic constraints for conversion of syngas to biofuels in acetogenic bacteria [J]. Biotechnol Biofuels, 2015, 8: 210. |
| [27] | Najafpour G, Younesi H. Ethanol and acetate synthesis from waste gas using batch culture of Clostridium ljungdahlii [J]. Enzyme Microb Technol, 2006, 38(1/2): 223-228. |
| [28] | Kerby RL, Ludden PW, Roberts GP. Carbon monoxide-dependent growth of Rhodospirillum rubrum [J]. J Bacteriol, 1995, 177(8): 2241-2244. |
| [29] | Kim MS, Bae SS, Kim YJ, et al. CO-dependent H2 production by genetically engineered Thermococcus onnurineus NA1 [J]. Appl Environ Microbiol, 2013, 79(6): 2048-2053. |
| [30] | Utturkar SM, Klingeman DM, Bruno-Barcena JM, et al. Sequence data for Clostridium autoethanogenum using three generations of sequencing technologies [J]. Sci Data, 2015, 2: 150014. |
| [31] | Dong HJ, Zhang YP, Dai ZJ, et al. Engineering Clostridium strain to accept unmethylated DNA [J]. PLoS One, 2010, 5(2): e9038. |
| [32] | Cui GZ, Hong W, Zhang J, et al. Targeted gene engineering in Clostridium cellulolyticum H10 without methylation [J]. J Microbiol Meth, 2012, 89(3): 201-208. |
| [33] | Heap JT, Pennington OJ, Cartman ST, et al. A modular system for Clostridium shuttle plasmids [J]. J Microbiol Meth, 2009, 78(1): 79-85. |
| [34] | Molitor B, Kirchner K, Henrich AW, et al. Expanding the molecular toolkit for the homoacetogen Clostridium ljungdahlii [J]. Sci Rep, 2016, 6: 31518. |
| [35] | Charubin K, Hill JD, Papoutsakis ET. DNA transfer between two different species mediated by heterologous cell fusion in Clostridium coculture [J]. mBio, 2024, 15(2): e0313323. |
| [36] | Purdy D, O'Keeffe TAT, Elmore M, et al. Conjugative transfer of clostridial shuttle vectors from Escherichia coli to Clostridium difficile through circumvention of the restriction barrier [J]. Mol Microbiol, 2002, 46(2): 439-452. |
| [37] | Desai RP, Papoutsakis ET. Antisense RNA strategies for metabolic engineering of Clostridium acetobutylicum [J]. Appl Environ Microbiol, 1999, 65(3): 936-945. |
| [38] | Perret S, Maamar H, Bélaich JP, et al. Use of antisense RNA to modify the composition of cellulosomes produced by Clostridium cellulolyticum [J]. Mol Microbiol, 2004, 51(2): 599-607. |
| [39] | Raju D, Setlow P, Sarker MR. Antisense-RNA-mediated decreased synthesis of small, acid-soluble spore proteins leads to decreased resistance of Clostridium perfringens spores to moist heat and UV radiation [J]. Appl Environ Microbiol, 2007, 73(7): 2048-2053. |
| [40] | Shao LJ, Hu SY, Yang Y, et al. Targeted gene disruption by use of a group II intron (targetron) vector in Clostridium acetobutylicum [J]. Cell Res, 2007, 17(11): 963-965. |
| [41] | Yoo M, Croux C, Meynial-Salles I, et al. Elucidation of the roles of adhE1 and adhE2 in the primary metabolism of Clostridium acetobutylicum by combining in-frame gene deletion and a quantitative system-scale approach [J]. Biotechnol Biofuels, 2016, 9: 92. |
| [42] | Cooksley CM, Zhang Y, Wang HZ, et al. Targeted mutagenesis of the Clostridium acetobutylicum acetone-butanol-ethanol fermentation pathway [J]. Metab Eng, 2012, 14(6): 630-641. |
| [43] | Leang C, Ueki T, Nevin KP, et al. A genetic system for Clostridium ljungdahlii: a chassis for autotrophic production of biocommodities and a model homoacetogen [J]. Appl Environ Microbiol, 2013, 79(4): 1102-1109. |
| [44] | Croux C, Nguyen NPT, Lee J, et al. Construction of a restriction-less, marker-less mutant useful for functional genomic and metabolic engineering of the biofuel producer Clostridium acetobutylicum [J]. Biotechnol Biofuels, 2016, 9: 23. |
| [45] | Al-Hinai MA, Fast AG, Papoutsakis ET. Novel system for efficient isolation of Clostridium double-crossover allelic exchange mutants enabling markerless chromosomal gene deletions and DNA integration [J]. Appl Environ Microbiol, 2012, 78(22): 8112-8121. |
| [46] | Cartman ST, Kelly ML, Heeg D, et al. Precise manipulation of the Clostridium difficile chromosome reveals a lack of association between the tcdC genotype and toxin production [J]. Appl Environ Microbiol, 2012, 78(13): 4683-4690. |
| [47] | Ng YK, Ehsaan M, Philip S, et al. Expanding the repertoire of gene tools for precise manipulation of the Clostridium difficile genome: allelic exchange using pyrE alleles [J]. PLoS One, 2013, 8(2): e56051. |
| [48] | Nariya H, Miyata S, Suzuki M, et al. Development and application of a method for counterselectable in-frame deletion in Clostridium perfringens [J]. Appl Environ Microbiol, 2011, 77(4): 1375-1382. |
| [49] | Ueki T, Nevin KP, Woodard TL, et al. Converting carbon dioxide to butyrate with an engineered strain of Clostridium ljungdahlii [J]. mBio, 2014, 5(5): e01636-14. |
| [50] | 杨君仪, 鲍江舰, 邵瑞瑞, 等. CD630_27900基因缺失显著降低艰难拟梭菌自溶速率、毒力及对酸和抗生素的耐受性 [J]. 微生物学报, 2023. 63(6): 2440-2455. |
| Yang JY, Bao JJ, Shao RR, et al. CD630_27900 gene deletion significantly reduces autolysis rate and virulence of Clostridioides difficile and the tolerance to acids and antibiotics [J]. Acta Microbiologica Sinica, 2023. 63(6): 2440-2455. | |
| [51] | 鲍江舰, 杨君仪, 邵瑞瑞, 等. fliL基因显著影响艰难拟梭菌运动功能及产孢能力 [J]. 生物工程学报, 2023. 39(4): 1578-1595. |
| Bao JJ, Yang JY, Shao RR, et al. The fliL gene significantly affects the motility and sporulation abilities of Clostridioides difficile [J]. Chinese Journal of Biotechnology, 2023. 39(4): 1578-1595. | |
| [52] | Ehsaan M, Kuehne SA, Minton NP. Clostridium difficile genome editing using pyrE alleles [J]. Methods Mol Biol, 2016, 1476: 35-52. |
| [53] | Annan FJ, Al-Sinawi B, Humphreys CM, et al. Engineering of vitamin prototrophy in Clostridium ljungdahlii and Clostridium autoethanogenum [J]. Appl Microbiol Biotechnol, 2019, 103(11): 4633-4648. |
| [54] | Kovács K, Willson BJ, Schwarz K, et al. Secretion and assembly of functional mini-cellulosomes from synthetic chromosomal operons in Clostridium acetobutylicum ATCC 824 [J]. Biotechnol Biofuels, 2013, 6(1): 117. |
| [55] | Huang H, Chai CS, Li N, et al. CRISPR/Cas9-based efficient genome editing in Clostridium ljungdahlii, an autotrophic gas-fermenting bacterium [J]. ACS Synth Biol, 2016, 5(12): 1355-1361. |
| [56] | Xu T, Li YC, Shi Z, et al. Efficient genome editing in Clostridium cellulolyticum via CRISPR-Cas9 nickase [J]. Appl Environ Microbiol, 2015, 81(13): 4423-4431. |
| [57] | Komor AC, Kim YB, Packer MS, et al. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage [J]. Nature, 2016, 533(7603): 420-424. |
| [58] | Kim YB, Komor AC, Levy JM, et al. Increasing the genome-targeting scope and precision of base editing with engineered Cas9-cytidine deaminase fusions [J]. Nat Biotechnol, 2017, 35(4): 371-376. |
| [59] | Gaudelli NM, Komor AC, Rees HA, et al. Programmable base editing of A·T to G·C in genomic DNA without DNA cleavage [J]. Nature, 2017, 551(7681): 464-471. |
| [60] | Zhao R, Liu YQ, Zhang H, et al. CRISPR-Cas12a-mediated gene deletion and regulation in Clostridium ljungdahlii and its application in carbon flux redirection in synthesis gas fermentation [J]. ACS Synth Biol, 2019, 8(10): 2270-2279. |
| [61] | Banerjee A, Leang C, Ueki T, et al. Lactose-inducible system for metabolic engineering of Clostridium ljungdahlii [J]. Appl Environ Microbiol, 2014, 80(8): 2410-2416. |
| [62] | Woolston BM, Emerson DF, Currie DH, et al. Rediverting carbon flux in Clostridium ljungdahlii using CRISPR interference (CRISPRi) [J]. Metab Eng, 2018, 48: 243-253. |
| [63] | Yang GH, Jia DC, Jin L, et al. Rapid generation of universal synthetic promoters for controlled gene expression in both gas-fermenting and saccharolytic Clostridium species [J]. ACS Synth Biol, 2017, 6(9): 1672-1678. |
| [64] | Girbal L, Mortier-Barrière I, Raynaud F, et al. Development of a sensitive gene expression reporter system and an inducible promoter-repressor system for Clostridium acetobutylicum [J]. Appl Environ Microbiol, 2003, 69(8): 4985-4988. |
| [65] | Dong HJ, Tao WW, Zhang YP, et al. Development of an anhydrotetracycline-inducible gene expression system for solvent-producing Clostridium acetobutylicum: a useful tool for strain engineering [J]. Metab Eng, 2012, 14(1): 59-67. |
| [66] | Heap JT, Pennington OJ, Cartman ST, et al. The ClosTron: a universal gene knock-out system for the genus Clostridium [J]. J Microbiol Meth, 2007, 70(3): 452-464. |
| [67] | Zhang Y, Grosse-Honebrink A, Minton NP. A universal mariner transposon system for forward genetic studies in the genus Clostridium [J]. PLoS One, 2015, 10(4): e0122411. |
| [68] | Drepper T, Huber R, Heck A, et al. Flavin mononucleotide-based fluorescent reporter proteins outperform green fluorescent protein-like proteins as quantitative in vivo real-time reporters [J]. Appl Environ Microbiol, 2010, 76(17): 5990-5994. |
| [69] | Drepper T, Eggert T, Circolone F, et al. Reporter proteins for in vivo fluorescence without oxygen [J]. Nat Biotechnol, 2007, 25(4): 443-445. |
| [70] | Mukherjee A, Schroeder CM. Flavin-based fluorescent proteins: emerging paradigms in biological imaging [J]. Curr Opin Biotechnol, 2015, 31: 16-23. |
| [71] | Whitham JM, Tirado-Acevedo O, Chinn MS, et al. Metabolic response of Clostridium ljungdahlii to oxygen exposure [J]. Appl Environ Microbiol, 2015, 81(24): 8379-8391. |
| [72] | Koepke M, Liew F. Genetically engineered bacterium with altered carbon monoxide dehydrogenase (CODH) activity: US9365873 [P]. 2016-06-14. |
| [73] | Han S, Gao XY, Ying HJ, et al. NADH gene manipulation for advancing bioelectricity in Clostridium ljungdahlii microbial fuel cells [J]. Green Chem, 2016, 18(8): 2473-2478. |
| [74] | Darmawi Juminaga, Arthur Shockley, and Robert Nogle. Recombinant Microorganisms and Uses Therefor[P]. 2022, WO2022170191A1. |
| [75] | Michael Koepke, Shilpa Nagaraju, and Chen Wendy. Recombinant Microorganisms and Methods of Use Thereof[P]. 2018, US 9057071B2. |
| [76] | Liew FE, Nogle R, Abdalla T, et al. Carbon-negative production of acetone and isopropanol by gas fermentation at industrial pilot scale [J]. Nat Biotechnol, 2022, 40(3): 335-344. |
| [77] | Vögeli B, Schulz L, Garg S, et al. Cell-free prototyping enables implementation of optimized reverse β-oxidation pathways in heterotrophic and autotrophic bacteria [J]. Nat Commun, 2022, 13(1): 3058. |
| [78] | Jin S, Bae JY, Song Y, et al. Synthetic biology on acetogenic bacteria for highly efficient conversion of C1 gases to biochemicals [J]. Int J Mol Sci, 2020, 21(20): 7639. |
| [79] | Jia DC, He MY, Tian Y, et al. Metabolic engineering of gas-fermenting Clostridium ljungdahlii for efficient co-production of isopropanol, 3-hydroxybutyrate, and ethanol [J]. ACS Synth Biol, 2021, 10(10): 2628-2638. |
| [80] | Schiel-Bengelsdorf B, Dürre P. Pathway engineering and synthetic biology using acetogens [J]. FEBS Lett, 2012, 586(15): 2191-2198. |
| [81] | Beck Zachary Q, Cervin Marguerite A, Chotani Gopal K, et al. Recombinant Anaerobic Acetogenic Bacteria for Production of Isoprene and/or Industrial Bio-Products Using Synthesis Gas: WO2014193473(A1) [P]. 2014-12-04. |
| [82] | Feng J, Zhang J, Ma YC, et al. Renewable fatty acid ester production in Clostridium [J]. Nat Commun, 2021, 12(1): 4368. |
| [83] | 刘昊鹏, 刘超, 王雯, 等. 基于厌氧微生物的碳链延长合成高价值化学品反应机理及研究进展:不同电子供体 [J]. 北京化工大学学报(自然科学版), 2020. 47(5): 1-17. |
| Liu HP, Liu C, Wang W, et al. Advances in understanding the mechanism of chain elongation with anaerobic microbes for the synthesis of high value-added chemicals: the effect of different electron donors [J]. Journal of Beijing University of Chemical Technology (Natural Science), 2020. 47(5): 1-17. |
| [1] | LU Tian-yi, LI Ai-peng, FEI Qiang. Research Progress in the Biosynthesis of Polylactic Acid [J]. Biotechnology Bulletin, 2025, 41(4): 47-60. |
| [2] | RAO Jun, ZHAO Chen, LI Duan-hua, LIAO Hao, HUANG Jia-yu, WANG Lu. Application of Auto-induction Strategy in Ergothioneine Biosynthesis [J]. Biotechnology Bulletin, 2025, 41(1): 333-346. |
| [3] | ZHANG Mei-yu, ZHAO Yu-bin, WANG Ling-yun, SONG Yuan-da, ZHAO Xin-he, REN Xiao-jie. Research Progress in the Production of Functional Fatty Acid DHA by Microalga Thraustochytrids [J]. Biotechnology Bulletin, 2024, 40(6): 81-94. |
| [4] | HE Si-cheng, ZHANG Zi-yuan, HAN Yu-qing, MIAO Lin, ZHANG Cui-ying, YU Ai-qun. Research Progress in the Production of Polyunsaturated Fatty Acids by Yarrowia lipolytica Cell Factories [J]. Biotechnology Bulletin, 2024, 40(1): 72-85. |
| [5] | LI Liang, XU Shan-shan, JIANG Yan-jun. Research Progress in the Production of Ergothioneine by Biosynthesis [J]. Biotechnology Bulletin, 2024, 40(1): 86-99. |
| [6] | XUE Ning, WANG Jin, LI Shi-xin, LIU Ye, CHENG Hai-jiao, ZHANG Yue, MAO Yu-feng, WANG Meng. Construction of L-phenylalanine High-producing Corynebacterium glutamicum Engineered Strains via Multi-gene Simultaneous Regulation Combined with High-throughput Screening [J]. Biotechnology Bulletin, 2023, 39(9): 268-280. |
| [7] | CHENG Ya-nan, ZHANG Wen-cong, ZHOU Yuan, SUN Xue, LI Yu, LI Qing-gang. Synthetic Pathway Construction of Producing 2'-fucosyllactose by Lactococcus lactis and Optimization of Fermentation Medium [J]. Biotechnology Bulletin, 2023, 39(9): 84-96. |
| [8] | ZHAO Si-jia, WANG Xiao-lu, SUN Ji-lu, TIAN Jian, ZHANG Jie. Modification of Pichia pastoris for Erythritol Production by Metabolic Engineering [J]. Biotechnology Bulletin, 2023, 39(8): 137-147. |
| [9] | LI Yu-zhen, MEI Tian-xiu, LI Zhi-wen, WANG Qi, LI Jun, ZOU Yue, ZHAO Xin-qing. Advances in Genomic Studies and Metabolic Engineering of Red Yeasts [J]. Biotechnology Bulletin, 2023, 39(7): 67-79. |
| [10] | YU Hui-li, LI Ai-tao. Application of Cytochrome P450 in the Biosynthesis of Flavors and Fragrances [J]. Biotechnology Bulletin, 2023, 39(4): 24-37. |
| [11] | LI Hai-ning, ZHANG Hong-bing, GENG Ge-xia, LI Ran, JIA Zhen-hua. Application and Biosynthesis Strategies of Unnatural Amino Acids [J]. Biotechnology Bulletin, 2023, 39(12): 43-55. |
| [12] | QIU Yi-bin, MA Yan-qin, SHA Yuan-yuan, ZHU Yi-fan, SU Er-zheng, LEI Peng, LI Sha, XU Hong. Research Progress in Molecular Genetic Manipulation Technology of Bacillus amyloliquefaciens and Its Application [J]. Biotechnology Bulletin, 2022, 38(2): 205-217. |
| [13] | MA Yan-qin, QIU Yi-bin, LI Sha, XU Hong. Research Progress in the Biosynthesis and Metabolic Engineering of Hyaluronic Acid [J]. Biotechnology Bulletin, 2022, 38(2): 252-262. |
| [14] | YE Jian-wen, CHEN Jiang-nan, ZHANG Xu, Wu Fu-qing, CHEN Guo-qiang. Dynamic Control:An Efficient Strategy for Metabolically Engineering Microbial Cell Factories [J]. Biotechnology Bulletin, 2020, 36(6): 1-12. |
| [15] | WANG Ke-wen ,YIN Xue, WANG Yu ,LI Yu-hua. Application of Selection and Optimization of Promoter in Metabolic Engineering of Saccharomyces cerevisiae [J]. Biotechnology Bulletin, 2018, 34(6): 38-47. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||