








Biotechnology Bulletin ›› 2025, Vol. 41 ›› Issue (10): 303-312.doi: 10.13560/j.cnki.biotech.bull.1985.2025-0488
Previous Articles Next Articles
LIANG Xin-min1( ), CUI Yu-qin1,2, LEI Meng-ting1,3, HAN Jing1, JIA Ding-hong1, WANG Bo1, PENG Wei-hong1,2, HE Xiao-lan1,3, LIU Xun1(
), CUI Yu-qin1,2, LEI Meng-ting1,3, HAN Jing1, JIA Ding-hong1, WANG Bo1, PENG Wei-hong1,2, HE Xiao-lan1,3, LIU Xun1( )
)
Received:2025-05-13
Online:2025-10-26
Published:2025-10-28
Contact:
LIU Xun
E-mail:liangxinmin_lxm@163.com;liuxun0712@163.com
LIANG Xin-min, CUI Yu-qin, LEI Meng-ting, HAN Jing, JIA Ding-hong, WANG Bo, PENG Wei-hong, HE Xiao-lan, LIU Xun. Expression Patterns of Tyrosinase and Laccase Genes in Flammulina filiformis with Different Colors[J]. Biotechnology Bulletin, 2025, 41(10): 303-312.
基因 Gene | 正向引物 Forward primer (5′-3′) | 反向引物 Reverse primer (5′-3′) |
|---|---|---|
| GAPDH | CCTCTGCTCACTTGAAGGGT | GCGTTGGAGATGACTTTGAA |
| TYR1 | AGGTGTGGTGCATCCAGAAG | GAACCAGAAGGACTTCGCCA |
| TYR2 | CGACCGCATGCTCTCACTAT | GGTGTCAACGGGGTATTGGT |
| TYR3 | GTACTGGAACGAACTCCCCG | CTTCCCCTGGGCCTAAGTTG |
| Lac1 | GGCCATTATGTCGACGGTCT | ACCAATCGCCAAGAACGACT |
| Lac2 | TCTACGATCCCGACGATCCA | GGCCGTTGATCAGGATAGCA |
| Lac3 | GTTTGCGAAGACTTTGGCGT | GACATGCTGACCAGACGGAA |
| Lac4 | ACCTGCGACGTTGAGAATGT | TGGGGTGCTCATCAATGGAC |
| Lac5 | CATCCATCGGCCTAACCGAA | TTTTCCCGCGACTACGTTGA |
| Lac6 | AGTGCAAGGAACTCGCTACC | TGAGTGTTGATCCCATCGGC |
| Lac7 | TTGTCCATGCCAATCAGCCT | GCGTCAGGAGCACCTTCATA |
| Lac8 | CGCTACCGCTTCCGGATTAT | TGGCTGCACGTTGATACCAT |
| Lac9 | CCAACGCTACTCCCTCATCC | ACATAATGCAGCACACCCGA |
| Lac10 | ATCTCCTGCGACCCAAACTG | ACTGAATCGACGAGAAGCGG |
| Lac11 | GGACCGACATCGAAGAGTCC | GCCCAGTTTGCGTTGGTATC |
Table 1 Primers used in this study
基因 Gene | 正向引物 Forward primer (5′-3′) | 反向引物 Reverse primer (5′-3′) |
|---|---|---|
| GAPDH | CCTCTGCTCACTTGAAGGGT | GCGTTGGAGATGACTTTGAA |
| TYR1 | AGGTGTGGTGCATCCAGAAG | GAACCAGAAGGACTTCGCCA |
| TYR2 | CGACCGCATGCTCTCACTAT | GGTGTCAACGGGGTATTGGT |
| TYR3 | GTACTGGAACGAACTCCCCG | CTTCCCCTGGGCCTAAGTTG |
| Lac1 | GGCCATTATGTCGACGGTCT | ACCAATCGCCAAGAACGACT |
| Lac2 | TCTACGATCCCGACGATCCA | GGCCGTTGATCAGGATAGCA |
| Lac3 | GTTTGCGAAGACTTTGGCGT | GACATGCTGACCAGACGGAA |
| Lac4 | ACCTGCGACGTTGAGAATGT | TGGGGTGCTCATCAATGGAC |
| Lac5 | CATCCATCGGCCTAACCGAA | TTTTCCCGCGACTACGTTGA |
| Lac6 | AGTGCAAGGAACTCGCTACC | TGAGTGTTGATCCCATCGGC |
| Lac7 | TTGTCCATGCCAATCAGCCT | GCGTCAGGAGCACCTTCATA |
| Lac8 | CGCTACCGCTTCCGGATTAT | TGGCTGCACGTTGATACCAT |
| Lac9 | CCAACGCTACTCCCTCATCC | ACATAATGCAGCACACCCGA |
| Lac10 | ATCTCCTGCGACCCAAACTG | ACTGAATCGACGAGAAGCGG |
| Lac11 | GGACCGACATCGAAGAGTCC | GCCCAGTTTGCGTTGGTATC |
基因名 Gene name | 蛋白大小Protein size (aa) | 分子量 Molecular weight (kD) | 等电点 pI | 不稳定指数Instability index | 亲水性平均值 Grand average of hydropathicity | 亚细胞定位 Subcellular localization |
|---|---|---|---|---|---|---|
| TYR1 | 318 | 35.4 | 4.86 | 41.54 | -0.361 | Extracellular space |
| TYR2 | 336 | 37.2 | 6.22 | 37.63 | -0.369 | Endomembrane system |
| TYR3 | 311 | 35.2 | 5.11 | 31.54 | -0.292 | Extracellular space |
| Lac1 | 387 | 42.5 | 4.53 | 33.57 | -0.033 | Extracellular space |
| Lac2 | 441 | 47.3 | 4.57 | 33.28 | 0.135 | Plasma membrane |
| Lac3 | 516 | 55.8 | 4.18 | 30.58 | -0.024 | Extracellular space |
| Lac4 | 642 | 68.9 | 4.75 | 35.55 | 0.018 | Extracellular space |
| Lac5 | 398 | 44.4 | 4.91 | 30.93 | -0.299 | Extracellular space |
| Lac6 | 788 | 86.5 | 5.36 | 34.82 | -0.122 | Extracellular space |
| Lac7 | 446 | 49.3 | 5.02 | 25.42 | -0.204 | Extracellular space |
| Lac8 | 523 | 57.1 | 5.17 | 29.37 | -0.020 | Extracellular space |
| Lac9 | 371 | 40.5 | 5.11 | 38.78 | -0.029 | Cytoplasm |
| Lac10 | 391 | 43.2 | 5.82 | 37.78 | 0.096 | Extracellular space |
| Lac11 | 196 | 21.4 | 4.17 | 27.25 | 0.051 | Extracellular space |
Table 2 Physicochemical properties of tyrosinase and laccase family members in F. filiformis
基因名 Gene name | 蛋白大小Protein size (aa) | 分子量 Molecular weight (kD) | 等电点 pI | 不稳定指数Instability index | 亲水性平均值 Grand average of hydropathicity | 亚细胞定位 Subcellular localization |
|---|---|---|---|---|---|---|
| TYR1 | 318 | 35.4 | 4.86 | 41.54 | -0.361 | Extracellular space |
| TYR2 | 336 | 37.2 | 6.22 | 37.63 | -0.369 | Endomembrane system |
| TYR3 | 311 | 35.2 | 5.11 | 31.54 | -0.292 | Extracellular space |
| Lac1 | 387 | 42.5 | 4.53 | 33.57 | -0.033 | Extracellular space |
| Lac2 | 441 | 47.3 | 4.57 | 33.28 | 0.135 | Plasma membrane |
| Lac3 | 516 | 55.8 | 4.18 | 30.58 | -0.024 | Extracellular space |
| Lac4 | 642 | 68.9 | 4.75 | 35.55 | 0.018 | Extracellular space |
| Lac5 | 398 | 44.4 | 4.91 | 30.93 | -0.299 | Extracellular space |
| Lac6 | 788 | 86.5 | 5.36 | 34.82 | -0.122 | Extracellular space |
| Lac7 | 446 | 49.3 | 5.02 | 25.42 | -0.204 | Extracellular space |
| Lac8 | 523 | 57.1 | 5.17 | 29.37 | -0.020 | Extracellular space |
| Lac9 | 371 | 40.5 | 5.11 | 38.78 | -0.029 | Cytoplasm |
| Lac10 | 391 | 43.2 | 5.82 | 37.78 | 0.096 | Extracellular space |
| Lac11 | 196 | 21.4 | 4.17 | 27.25 | 0.051 | Extracellular space |
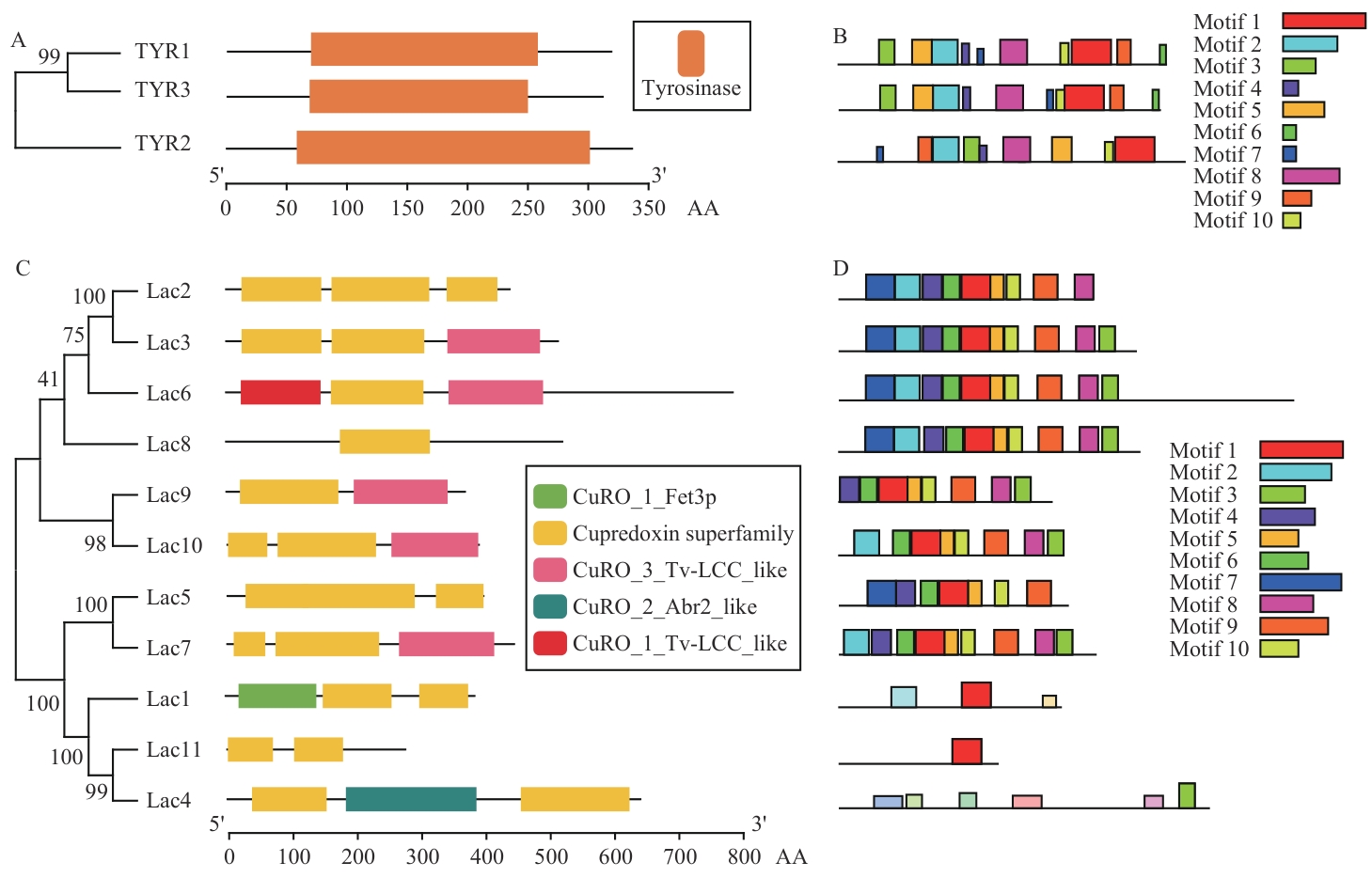
Fig. 3 Conserved domains and conserved motifs of the TYR and Lac family proteinsA: Conserved domain of TYR protein. B: Conserved motif of TYR protein. C: Conserved domain of Lac protein. D: Conserved motif of Lac protein
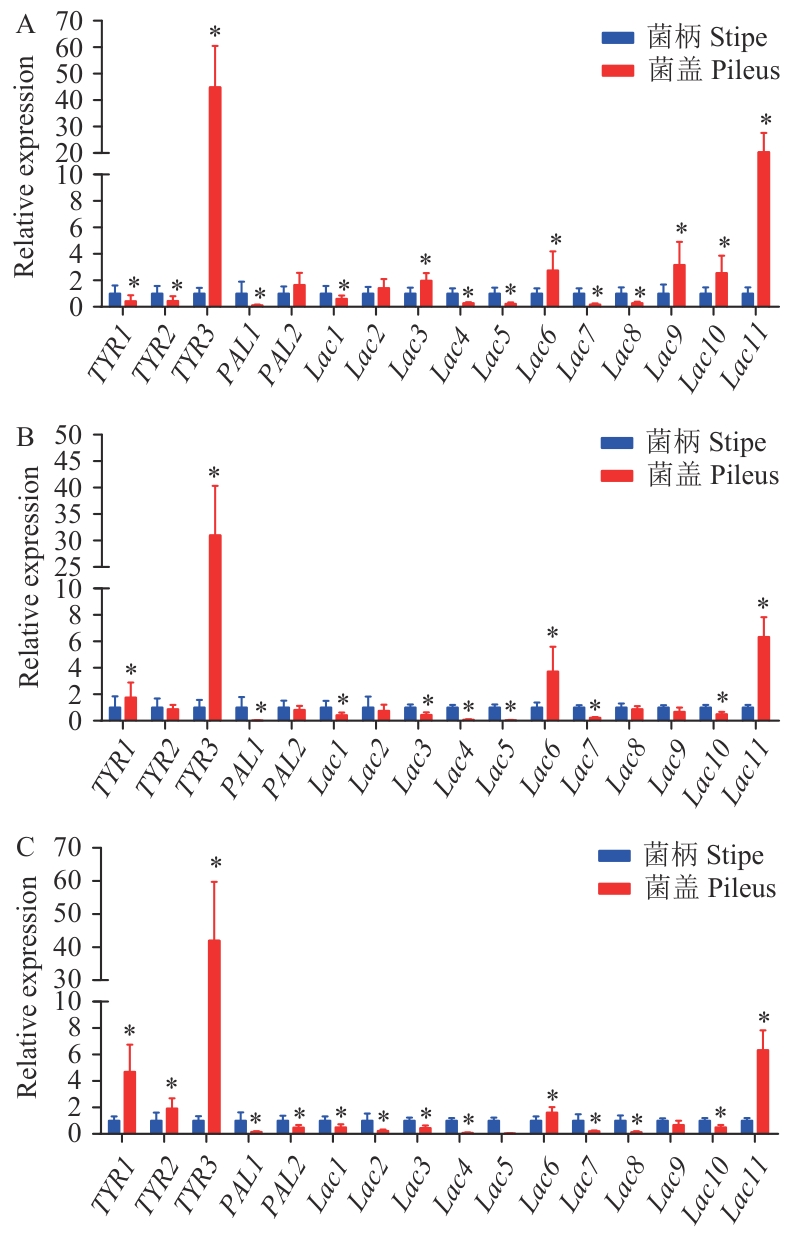
Fig. 4 Expression patterns of TYR and Lac genes in the epidermis of different-color F. filiformis stipes and pileusA: White F. filiformis. B: Yellow F. filiformis. C: Brown F. filiformis. *P<0.05
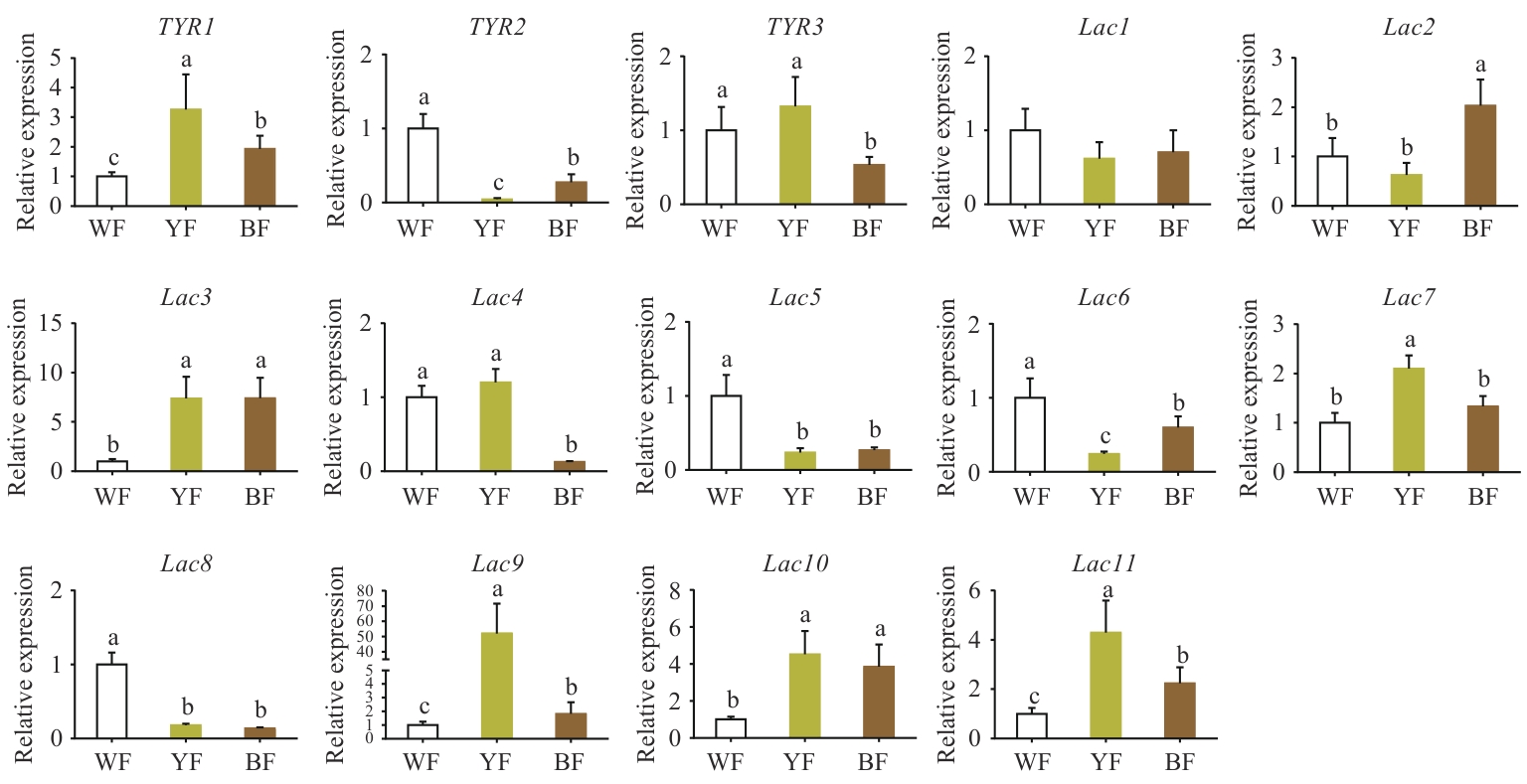
Fig. 5 Comparison of the relative expressions of TYR and Lac genes in the epidermis of pileus with different colorsWF: White pileus epidermal tissue. YF: Yellow pileus epidermal tissue. BF: Brown pileus epidermal tissue. Different letters refer to significant difference (P<0.05). The same below
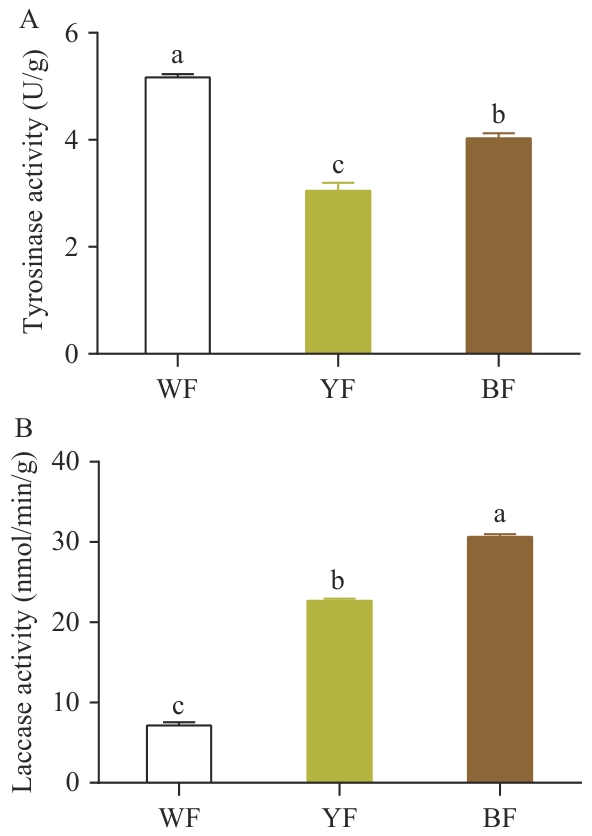
Fig. 6 Comparative analysis of tyrosinase and laccase activities in the epidermis of pileus with different colorsA: Tyrosinase activity in the epidermal tissue of the pileus. B: Laccase activity in the epidermal tissue of the pileus
| [1] | Wang PM, Liu XB, Dai YC, et al. Phylogeny and species delimitation of Flammulina: taxonomic status of winter mushroom in east Asia and a new European species identified using an integrated approach [J]. Mycol Prog, 2018, 17(9): 1013-1030. |
| [2] | 戴玉成, 杨祝良. 中国五种重要食用菌学名新注 [J]. 菌物学报, 2018, 37(12): 1572-1577. |
| Dai YC, Yang ZL. Notes on the nomenclature of five important edible fungi in China [J]. Mycosystema, 2018, 37(12): 1572-1577. | |
| [3] | 中国食用菌协会. 2023年度全国食用菌统计调查结果分析[J]. 中国食用菌, 2025, 44(1): 120-129. |
| China Association of Edible Fungi. Analysis of the results of the national edible fungi statistical survey in 2023[J]. Edible Fungi of China, 2025, 44(1): 120-129. | |
| [4] | 王波. 金针菇新品种'川金33'的选育和栽培应用 [J]. 食药用菌, 2018, 26(1): 38-39. |
| Wang B. Breeding and cultivation application of a new Flammulina velutipes variety ‘Chuanjin 33’ [J]. Edible Med Mushrooms, 2018, 26(1): 38-39. | |
| [5] | 刘询, 何晓兰, 贾定洪, 等. 基于非靶向代谢组学分析黄色和白色金针菇代谢物差异[J]. 菌物学报, 2024, 43(8): 73-85. |
| Liu X, He XL, Jia DH, et al. Analysis of metabolite difference between yellow and white strains of Flammulina filiformis based on untargeted metabolomics [J]. Mycosystema, 2024. 43(8): 73-85. | |
| [6] | Jolivet S, Arpin N, Wichers HJ, et al. Agaricus bisporus browning: a review [J]. Mycol Res, 1998, 102(12): 1459-1483. |
| [7] | Claus H, Decker H. Bacterial tyrosinases [J]. Syst Appl Microbiol, 2006, 29(1): 3-14. |
| [8] | 韩建荣. 木耳子实体阶段酪氨酸酶活性与温度光照和子实体颜色的关系 [J]. 中国食用菌, 1993, 12(2): 11-13. |
| Han JR. Relationship of activities of tyrosinase with the cultivation temperature and licht as well as the fruitbody colqur during fruiting of au ricul Aria au Ricula [J]. Edible Fungi China, 1993, 12(2): 11-13. | |
| [9] | Zhang Y, Wu XL, Huang CY, et al. Isolation and identification of pigments from oyster mushrooms with black, yellow and pink caps [J]. Food Chem, 2022, 372: 131171. |
| [10] | Zhang Y, Huang CY, van Peer AF, et al. Fine mapping and functional analysis of the gene PcTYR, involved in control of cap color of Pleurotus cornucopiae [J]. Appl Environ Microbiol, 2022, 88(7): e0217321. |
| [11] | 刘丽娜. 金针菇颜色突变型的遗传属性研究 [D]. 长沙: 湖南师范大学, 2016. |
| Liu LN. Study on genetic properties of color mutant of Flammulina velutipes [D]. Changsha: Hunan Normal University, 2016. | |
| [12] | Zeng JY, Shi DD, Chen Y, et al. FvbHLH1 regulates the accumulation of phenolic compounds in the yellow cap of Flammulina velutipes [J]. J Fungi, 2023, 9(11): 1063. |
| [13] | Im JH, Yu HW, Park CH, et al. Phenylalanine ammonia-lyase: a key gene for color discrimination of edible mushroom Flammulina velutipes [J]. J Fungi, 2023, 9(3): 339. |
| [14] | Liang YQ, Luo KM, Wang BL, et al. Inhibition of polyphenol oxidase for preventing browning in edible mushrooms: a review [J]. J Food Sci, 2024, 89(11): 6796-6817. |
| [15] | Martin E, Dubessay P, Record E, et al. Recent advances in laccase activity assays: a crucial challenge for applications on complex substrates [J]. Enzyme Microb Technol, 2024, 173: 110373. |
| [16] | Yan LL, Xu RP, Bian YB, et al. Expression profile of laccase gene family in white-rot basidiomycete Lentinula edodes under different environmental stresses [J]. Genes, 2019, 10(12): 1045. |
| [17] | Zhang JL, Tanaka Y, Ono A, et al. Gene expression analysis for stem browning in the mushroom Lentinula edodes [J]. Mycoscience, 2024, 65(5): 253-259. |
| [18] | 李子豪, 李小凤, 杜芳, 等. 刺芹侧耳不同生长发育阶段漆酶基因家族表达 [J]. 食用菌学报, 2021, 28(4): 1-6. |
| Li ZH, Li XF, Du F, et al. Expression of laccase gene family at different developmental stages of Pleurotus eryngii [J]. Acta Edulis Fungi, 2021, 28(4): 1-6. | |
| [19] | 刘冬梅, 刁文彤, 孙雪言, 等. 灵芝漆酶基因家族的克隆及其在不同发育阶段的表达分析 [J]. 微生物学杂志, 2025, 45(1): 34-42. |
| Liu DM, Diao WT, Sun XY, et al. Cloning and expression analysis of laccase genes family at developmental stages of Ganoderma lucidum [J]. Journal of Microbiology, 2025, 45(1): 34-42. | |
| [20] | He Y, Liu B, Ouyang XQ, et al. Whole-genome sequencing and fine map analysis of Pholiota nameko [J]. J Fungi, 2025, 11(2): 112. |
| [21] | 吉军, 张玉琎, 肖鑫, 等. 白假鬼伞漆酶基因家族鉴定及表达模式 [J]. 植物生理学报, 2023, 59(11): 2027-2038. |
| Ji J, Zhang YJ, Xiao X, et al. Identification and expression analysis on laccase gene family in Coprinellus disseminates [J]. Plant Physiology Journal, 2023, 59(11): 2027-2038. | |
| [22] | Liang XM, Han J, Cui YQ, et al. Whole-genome sequencing of Flammulina filiformis and multi-omics analysis in response to low temperature [J]. J Fungi, 2025, 11(3): 229. |
| [23] | 杜金茹, 王守现, 严冬, 等. 基于全基因组数据分析香菇黑色素合成途径及相关基因 [J]. 菌物学报, 2023, 42(5):1114-1128. |
| Du JR, Wang SX, Yan D, et al. Analyses of melanin synthesis pathway and the related genes in Lentinula edodes based on whole genome sequence comparison [J]. Mycosystema, 2023, 42(5): 1114-1128. | |
| [24] | Wilkins MR, Gasteiger E, Bairoch A, et al. Protein identification and analysis tools in the ExPASy server [J]. Methods Mol Biol, 1999, 112: 531-552. |
| [25] | Savojardo C, Martelli PL, Fariselli P, et al. BUSCA: an integrative web server to predict subcellular localization of proteins [J]. Nucleic Acids Res, 2018, 46(W1): W459-W466. |
| [26] | 王波, 刘询, 何晓兰, 等. 一株褐色金针菇及其应用:中国, CN202410393413.4 [P]. 2024-08-13. |
| Wang B, Liu X, He XL, et al. A brown Flammulina filiformis and its applications: China, CN202410393413.4 [P]. 2024-08-13. | |
| [27] | Yu CY. Molecular mechanism of manipulating seed coat coloration in oilseed Brassica species [J]. J Appl Genet, 2013, 54(2): 135-145. |
| [28] | Wang M, Song TT, Jin QL, et al. From white to reddish-brown: the anthocyanin journey in Stropharia rugosoannulata driven by auxin and genetic regulators [J]. J Agric Food Chem, 2025, 73(1): 954-966. |
| [29] | 吴林, 朱刚, 陈明杰, 等. 草菇基因组中11个漆酶同源基因的生物信息学分析及铜离子对其表达的影响 [J]. 菌物学报, 2014, 33(2): 323-333. |
| Wu L, Zhu G, Chen MJ, et al. The bioinformatic analyses and the gene expression induced by Cu2+ of 11 laccase homologous genes from Volvariella volvacea [J]. Mycosystema, 2014, 33(2): 323-333. | |
| [30] | 李雪松, 孙达锋, 华蓉, 等. 皱木耳漆酶生物信息学分析 [J]. 中国食用菌, 2023, 42(2): 52-59. |
| Li XS, Sun DF, Hua R, et al. Bioinformatics identification of laccase in Auricularia delicata [J]. Edible Fungi China, 2023, 42(2): 52-59. | |
| [31] | Fu NN, Li JX, Wang M, et al. Genes identification, molecular docking and dynamics simulation analysis of laccases from Amylostereum areolatum provides molecular basis of laccase bound to lignin [J]. Int J Mol Sci, 2020, 21(22): 8845. |
| [32] | Litvintseva AP, Henson JM. Cloning, characterization, and transcription of three laccase genes from Gaeumannomyces graminis var. tritici, the take-all fungus [J]. Appl Environ Microbiol, 2002, 68(3): 1305-1311. |
| [33] | Jin WS, Li JH, Feng HC, et al. Importance of a laccase gene (Lcc1) in the development of Ganoderma tsugae [J]. Int J Mol Sci, 2018, 19(2): 471. |
| [34] | Chen SC, Ge W, Buswell JA. Molecular cloning of a new laccase from the edible straw mushroom Volvariella volvacea: possible involvement in fruit body development [J]. FEMS Microbiol Lett, 2004, 230(2): 171-176. |
| [1] | ZHANG Chao-chao, HAN Kai-yuan, WANG Tong, CHEN Zhong. Cloning and Functional Analysis of PtoYABBY2 and PtoYABBY12 in Populus tomentosa [J]. Biotechnology Bulletin, 2025, 41(9): 256-264. |
| [2] | SHI Fa-chao, JIANG Yong-hua, LIU Hai-lun, WEN Ying-jie, YAN Qian. Cloning and Functional Analysis of LcTFL1 Gene in Litchi chinensis Sonn. [J]. Biotechnology Bulletin, 2025, 41(9): 159-167. |
| [3] | LIU Jia-li, SONG Jing-rong, ZHAO Wen-yu, ZHANG Xin-yuan, ZHAO Zi-yang, CAO Yi-bo, ZHANG Ling-yun. Identification of the R2R3-MYB Gene and Expression Analysis of Flavonoid Regulatory Genes in Blueberry [J]. Biotechnology Bulletin, 2025, 41(9): 124-138. |
| [4] | LI Yu-zhen, LI Meng-dan, ZHANG Wei, PENG Ting. Functional Study of RmEXPB2 Genein Rosa multiflora Based on the Identification of the Expansin Gene Family in Rosa sp. [J]. Biotechnology Bulletin, 2025, 41(9): 182-194. |
| [5] | HUANG Shi-yu, TIAN Shan-shan, YANG Tian-wei, GAO Man-rong, ZHANG Shang-wen. Genome-wide Identification and Expression Pattern Analysis of WRI1 Gene Family in Erythropalum scandens [J]. Biotechnology Bulletin, 2025, 41(8): 242-254. |
| [6] | KANG Qin, WANG Xia, SHEN Ming-yang, XU Jing-tian, CHEN Shi-lan, LIAO Ping-yang, XU Wen-zhi, WU Wei, XU Dong-bei. Cloning and Expression Analysis of the UV-B Receptor Gene McUVR8 in Mentha canadensis L. [J]. Biotechnology Bulletin, 2025, 41(8): 255-266. |
| [7] | DUAN Min-jie, LI Yi-fei, WANG Chun-ping, HUANG Ren-zhong, HUANG Qi-zhong, ZHANG Shi-cai. Association Analysis and Fingerprint Map Construction of Fruit Color Traits in Pepper via SSR Markers [J]. Biotechnology Bulletin, 2025, 41(7): 81-94. |
| [8] | WANG Fang, QIAO Shuai, SONG Wei, CUI Peng-juan, LIAO An-zhong, TAN Wen-fang, YANG Song-tao. Genome-wide Identification of the IbNRT2 Gene Family and Its Expression in Sweet Potato [J]. Biotechnology Bulletin, 2025, 41(7): 193-204. |
| [9] | XU Hui-zhen, SHANTWANA Ghimire, RAJU Kharel, YUE Yun, SI Huai-jun, TANG Xun. Analysis of the Potato SUMO E3 Ligase Gene Family and Cloning and Expression Pattern of StSIZ1 [J]. Biotechnology Bulletin, 2025, 41(6): 119-129. |
| [10] | WANG Tian-xi, YANG Bing-song, PAN Rong-jun, GAI Wen-xian, LIANG Mei-xia. Identification of the Apple PLATZ Gene Family and Functional Study of the MdPLATZ9 Gene [J]. Biotechnology Bulletin, 2025, 41(4): 176-187. |
| [11] | SONG Jia-yi, SU Yun-li, ZHENG Xing-yan, XIA Wen-nian, YANG Dong-mei, HU Hui-zhen. Identification of the Snapdragon Expansin Gene Family and Screening of Its Genes Related to Resistance to Sclerotinia sclerotiorum [J]. Biotechnology Bulletin, 2025, 41(4): 227-242. |
| [12] | FENG Xiao-kang, LIANG Qian, WANG Xue-feng, SUN Jie, XUE Fei. Identification of SEC1 Complex Components and Functional Validation of the GhSCY1 Gene in Cotton [J]. Biotechnology Bulletin, 2025, 41(3): 112-122. |
| [13] | SONG Shu-yi, JIANG Kai-xiu, LIU Huan-yan, HUANG Ya-cheng, LIU Lin-ya. Identification of the TCP Gene Family in Actinidia chinensis var. Hongyang and Their Expression Analysis in Fruit [J]. Biotechnology Bulletin, 2025, 41(3): 190-201. |
| [14] | KUANG Jian-hua, CHENG Zhi-peng, ZHAO Yong-jing, YANG Jie, CHEN Run-qiao, CHEN Long-qing, HU Hui-zhen. Expression Analysis of the GH3 Gene Family in Nelumbo nucifera underHormonal and Abiotic Stresses [J]. Biotechnology Bulletin, 2025, 41(2): 221-233. |
| [15] | QIAN Zheng-yi, WU Shao-fang, CAO Shu-yi, SONG Ya-xin, PAN Xin-feng, LI Zhao-wei, FAN Kai. Identification of the NAC Transcription Factors in Nymphaea colorata and Their Expression Analysis [J]. Biotechnology Bulletin, 2025, 41(2): 234-247. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||