








Biotechnology Bulletin ›› 2025, Vol. 41 ›› Issue (6): 344-354.doi: 10.13560/j.cnki.biotech.bull.1985.2024-1125
CHENG Hui-juan1( ), WANG Xin1, SHI Xiao-tao2, MA Dong-xu1,2(
), WANG Xin1, SHI Xiao-tao2, MA Dong-xu1,2( ), GONG Da-chun1(
), GONG Da-chun1( ), HU Jun-peng3, XIE Zhi-wen3
), HU Jun-peng3, XIE Zhi-wen3
Received:2024-12-17
Online:2025-06-26
Published:2025-06-30
Contact:
MA Dong-xu, GONG Da-chun
E-mail:1074063904@qq.com;18222712891@163.com;185195061@ qq.com
CHENG Hui-juan, WANG Xin, SHI Xiao-tao, MA Dong-xu, GONG Da-chun, HU Jun-peng, XIE Zhi-wen. Effects of Transcription Factor CREA Knockout on the Morphology and the Secretion of β-glucosidase in Aspergillus niger[J]. Biotechnology Bulletin, 2025, 41(6): 344-354.
| Name | Primer sequence(5′-3′) |
|---|---|
| 5srRNA-F | TACTGGGCCCGGGAAGATCTGGTTGGAGATTCCAGACTCAG |
| 5srRNA-R | CCTTGTCTGCTTACGCGATCACATACAACAGAAGGGATTC |
| sgRNA-F | GATCGCGTAAGCAGACAAGGGTTTTAGAGCTAGAAATAGC |
| sgRNA-R | CATGGTCATAGCTGTTTCCGAAAAAAGCACCGACTCGG |
| pFC332-F | ACCCTGTGAGTCTTGATAGAC |
| pFC332-R | CCGTTCGAGAGCATGATC |
| gtsy-F | ACTTATTACCCCGGGGTCCA |
| gtsy-R | CTGGACGGAGTAGAAGGGG |
| gtxy-F | TACACATGAATTTGATATAAAGGTCTGATGCC |
| gtxy-R | ACCCCTTCTACTCCGTCCAGGCGAATTCCGTTGCCGG |
| creAYZ-F | CAACATCACCCTTGCCCGAC |
| 18S-F | ACTCACCAGGTCCAGACAAAATAAG |
| 18S-R | AAGCAGACAAATCACTCCA |
| bglA-F | CAGGATGCCTCAGACTATCTTC |
| bglA-R | CGCCAAGAGAAACATACAGTTG |
| cbhA-F | CAGTTGTCATTGACTCGAACTG |
| cbhA-R | TTTCAGTGTCAATGAATCACCG |
| eglA-F | ACGATGTGCTCTCAATATGACA |
| eglA-R | ATCACTCACGAGCTTCTTGTTA |
Table 1 Primers used in this study
| Name | Primer sequence(5′-3′) |
|---|---|
| 5srRNA-F | TACTGGGCCCGGGAAGATCTGGTTGGAGATTCCAGACTCAG |
| 5srRNA-R | CCTTGTCTGCTTACGCGATCACATACAACAGAAGGGATTC |
| sgRNA-F | GATCGCGTAAGCAGACAAGGGTTTTAGAGCTAGAAATAGC |
| sgRNA-R | CATGGTCATAGCTGTTTCCGAAAAAAGCACCGACTCGG |
| pFC332-F | ACCCTGTGAGTCTTGATAGAC |
| pFC332-R | CCGTTCGAGAGCATGATC |
| gtsy-F | ACTTATTACCCCGGGGTCCA |
| gtsy-R | CTGGACGGAGTAGAAGGGG |
| gtxy-F | TACACATGAATTTGATATAAAGGTCTGATGCC |
| gtxy-R | ACCCCTTCTACTCCGTCCAGGCGAATTCCGTTGCCGG |
| creAYZ-F | CAACATCACCCTTGCCCGAC |
| 18S-F | ACTCACCAGGTCCAGACAAAATAAG |
| 18S-R | AAGCAGACAAATCACTCCA |
| bglA-F | CAGGATGCCTCAGACTATCTTC |
| bglA-R | CGCCAAGAGAAACATACAGTTG |
| cbhA-F | CAGTTGTCATTGACTCGAACTG |
| cbhA-R | TTTCAGTGTCAATGAATCACCG |
| eglA-F | ACGATGTGCTCTCAATATGACA |
| eglA-R | ATCACTCACGAGCTTCTTGTTA |
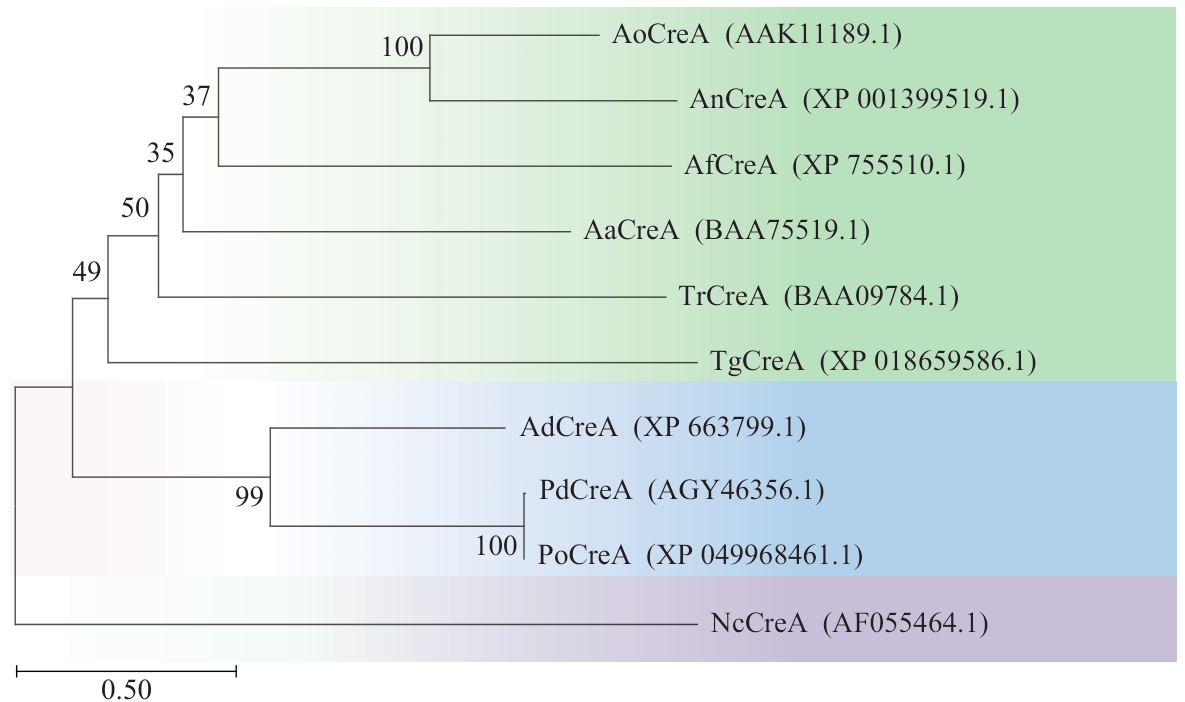
Fig. 1 Evolutionary tree analysis of CREA amino acid sequencesAoCreA: CREA in Aspergillus oryzae; AnCreA:CREA in Aspergillus niger; AfCreA: CREA in Aspergillus fumigatus; AaCreA: CREA in Aspergillus aculeatus; TrCreA: CREA in Trichoderma reesei; TgCreA: CREA in Trichoderma gamsii; AdCreA: CREA in Aspergillus nidulans; PdCreA: CREA in Penicillium decumens; PoCreA: CREA in Penicillium oxalicum; NcCreA: CREA in Neurospora crassa

Fig. 2 Construction of CRISPR-Cas9 targeting plasmid and donor DNAA: Construction of sgRNA expression cassette; B: construction of pFC332-target-sgRNA; C: construction of DNA donor; D-F: PCR detection of sgRNA expression cassette, pFC332-target-sgRNA and DNA donor, respectively
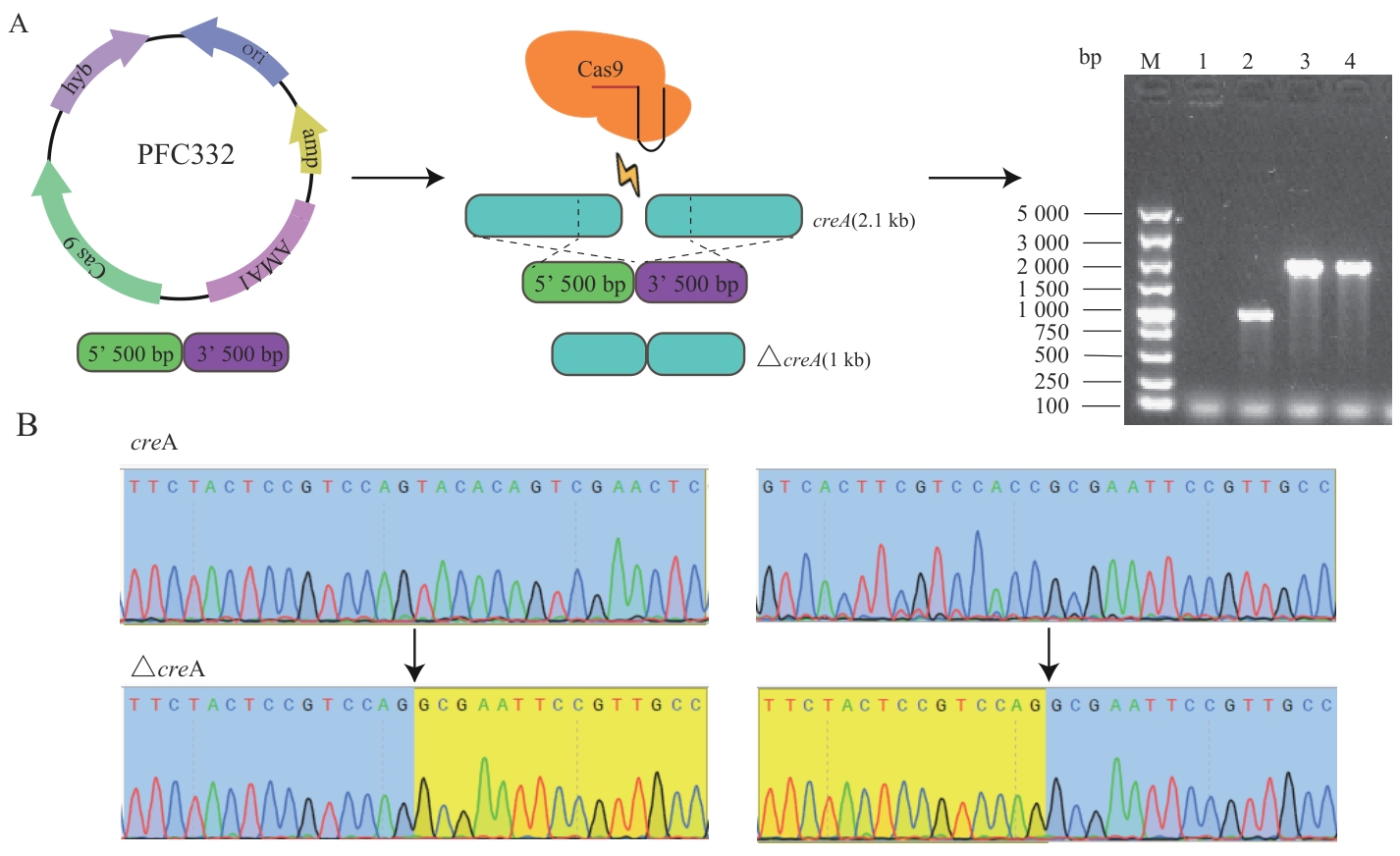
Fig. 3 Knockout of the creA gene in A. nigerA: Schematic diagram of creA gene editing and verification of PCR knockout; B:Sequencing imageof creA gene knockout

Fig. 4 Differences in the morphologies of A. niger in different mediaA: Cell morphology in PDA solid plates cultured for 48/60/72 h, respectively. B: Cell morphology in esculin solid plates cultured for 48/60/72 h, respectively. C: Cell morphology in liquid growth medium
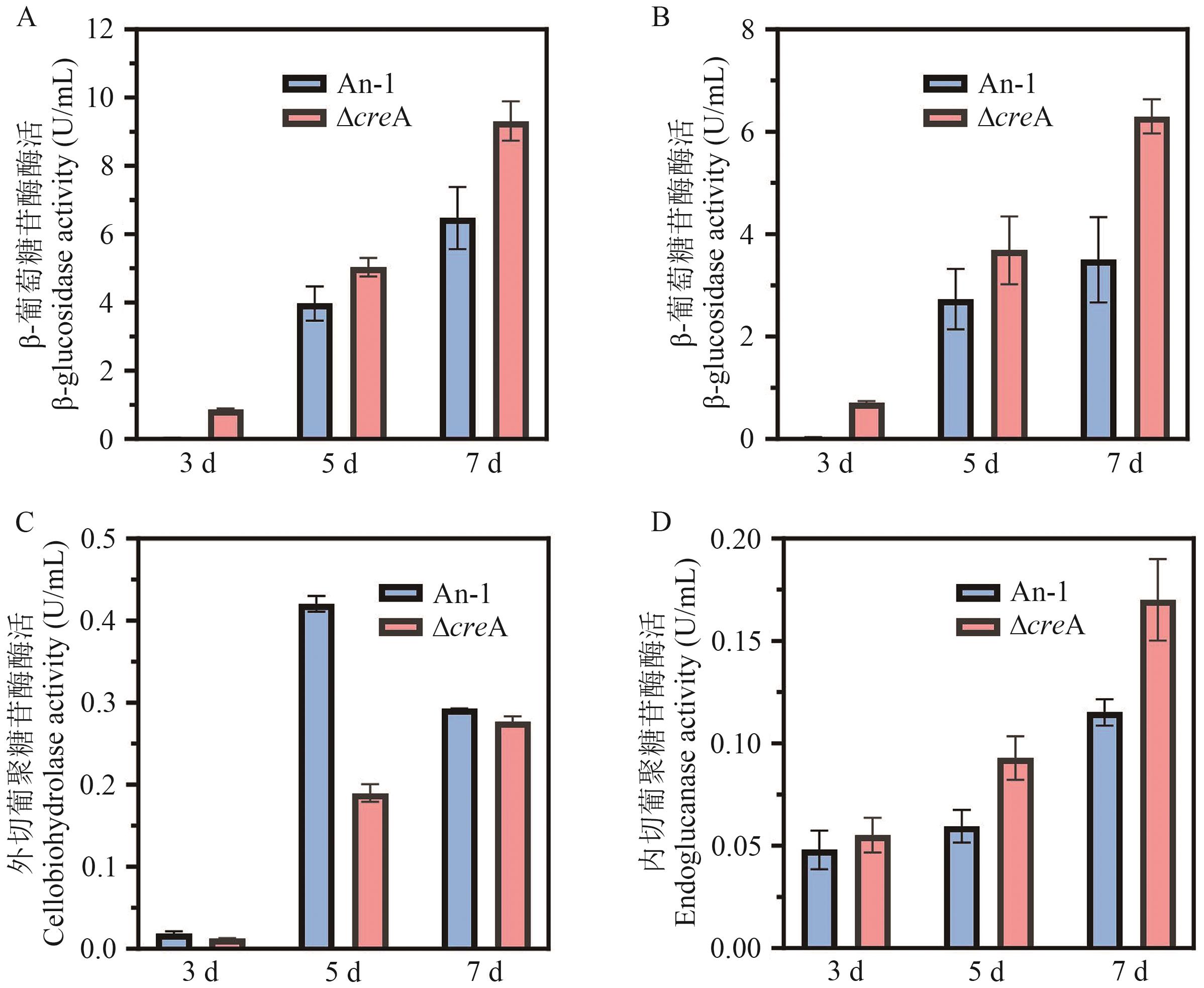
Fig. 5 Effects of creA knockout on the enzymatic activities of different enzymesA: BGL enzyme activity determined by cellobiose as substrate. B: BGL enzyme activity assayed with p-NPG as substrate. C: CBH enzyme activity assayed with p-NPC as substrate. D: EG enzyme activity assayed by CMC-Na as substrate

Fig. 6 Effects of creA knockout on expression and proteinA: Transcription levels of bglA, cbh1, and egl1 genes; B: SDS-PAGE on the 7 th day of fermentation, 1-3 refer to the fermented samples of △creA strain, and 4-6 refer to the fermentation samples of wild strains. C: Protein content and specific activity of BGL (cellobiose as substrate) on the fermentation of day 7
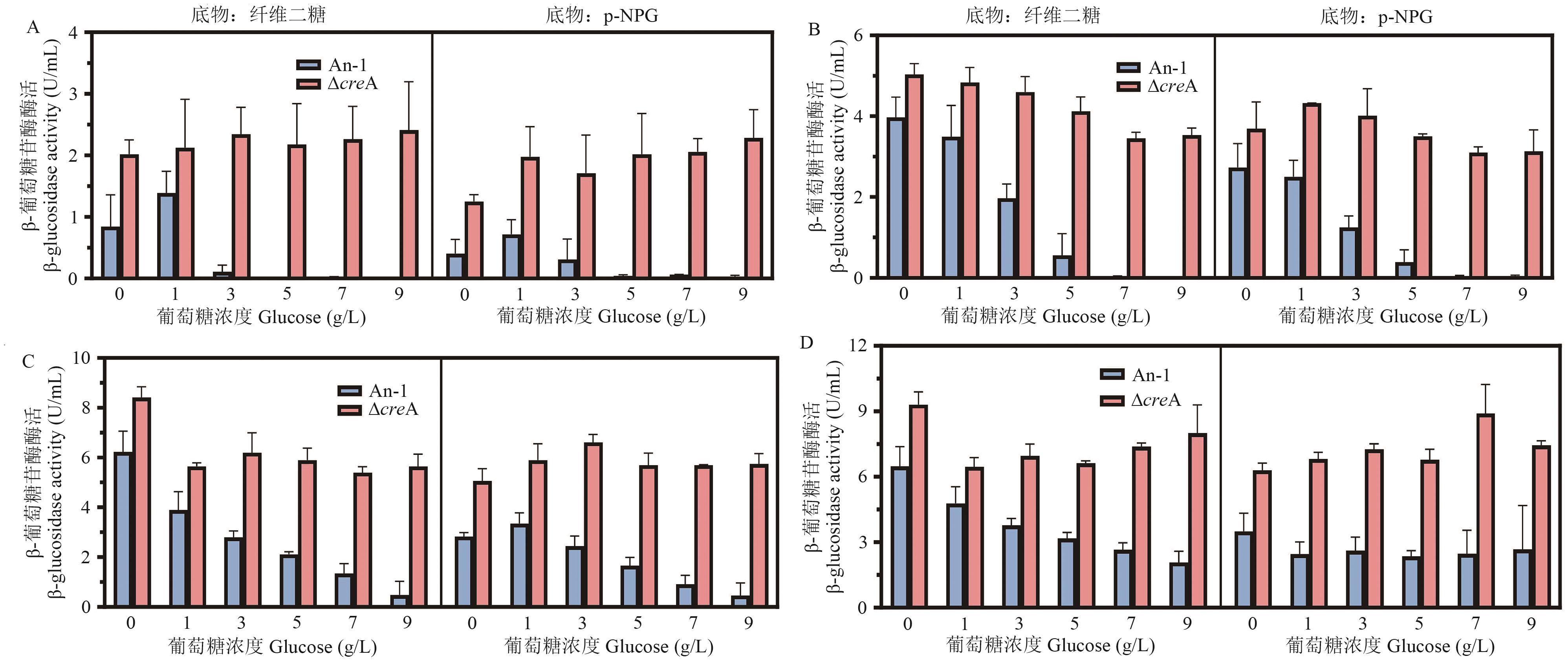
Fig. 7 Effects of creA knockout on BGL production by A. niger at different glucose concentrationsA: Enzyme activity level after 4 d of fermentation. B: Enzyme activity level after 5 d of fermentation. C: Enzyme activity level after 6 d of fermentation. D: Enzyme activity level for 7 d of fermentation
| 1 | Karimi K, Taherzadeh MJ. A critical review of analytical methods in pretreatment of lignocelluloses: Composition, imaging, and crystallinity [J]. Bioresour Technol, 2016, 200: 1008-1018. |
| 2 | Yamamoto K, Tamaru Y. Important roles of the cellulosome on degradation of plant biomass [M]// Gupta VK. New and future developments in microbial biotechnology and bioengineering. Amsterdam: Elsevier, 2016: 3-8. |
| 3 | de Paula RG, Antoniêto ACC, Ribeiro LFC, et al. Engineered microbial host selection for value-added bioproducts from lignocellulose [J]. Biotechnol Adv, 2019, 37(6): 107347. |
| 4 | Raj T, Chandrasekhar K, Naresh Kumar A, et al. Lignocellulosic biomass as renewable feedstock for biodegradable and recyclable plastics production: a sustainable approach [J]. Renew Sustain Energy Rev, 2022, 158: 112130. |
| 5 | Gao JQ, Yu W, Li YX, et al. Engineering co-utilization of glucose and xylose for chemical overproduction from lignocellulose [J]. Nat Chem Biol, 2023, 19(12): 1524-1531. |
| 6 | Singhania RR, Patel AK, Sukumaran RK, et al. Role and significance of beta-glucosidases in the hydrolysis of cellulose for bioethanol production [J]. Bioresour Technol, 2013, 127: 500-507. |
| 7 | Guo HL, Zhao Y, Chang JS, et al. Enzymes and enzymatic mechanisms in enzymatic degradation of lignocellulosic biomass: a mini-review [J]. Bioresour Technol, 2023, 367: 128252. |
| 8 | Ma XY, Li SP, Tong XX, et al. An overview on the current status and future prospects in Aspergillus cellulase production [J]. Environ Res, 2024, 244: 117866. |
| 9 | Sukumaran RK, Christopher M, Kooloth-Valappil P, et al. Addressing challenges in production of cellulases for biomass hydrolysis: Targeted interventions into the genetics of cellulase producing fungi [J]. Bioresour Technol, 2021, 329: 124746. |
| 10 | Wu VW, Thieme N, Huberman LB, et al. The regulatory and transcriptional landscape associated with carbon utilization in a filamentous fungus [J]. Proc Natl Acad Sci U S A, 2020, 117(11): 6003-6013. |
| 11 | Nakari-Setälä T, Paloheimo M, Kallio J, et al. Genetic modification of carbon catabolite repression in Trichoderma reesei for improved protein production [J]. Appl Environ Microbiol, 2009, 75(14): 4853-4860. |
| 12 | Yao GS, Li ZH, Gao LW, et al. Redesigning the regulatory pathway to enhance cellulase production in Penicillium oxalicum [J]. Biotechnol Biofuels, 2015, 8: 71. |
| 13 | 张飞, 白凤武, 赵心清. 丝状真菌纤维素酶合成诱导及转录调控 [J]. 生物工程学报, 2016, 32(11): 1481-1495. |
| Zhang F, Bai FW, Zhao XQ. Induction and regulation of cellulase expression in filamentous fungi: a review [J]. Chin J Biotechnol, 2016, 32(11): 1481-1495. | |
| 14 | Benocci T, Aguilar-Pontes MV, Zhou MM, et al. Regulators of plant biomass degradation in ascomycetous fungi [J]. Biotechnol Biofuels, 2017, 10: 152. |
| 15 | Bi FC, Barad S, Ment D, et al. Carbon regulation of environmental pH by secreted small molecules that modulate pathogenicity in phytopathogenic fungi [J]. Mol Plant Pathol, 2016, 17(8): 1178-1195. |
| 16 | Vautard-Mey G, Cotton P, Fèvre M. Expression and compartmentation of the glucose repressor CRE1 from the phytopathogenic fungus Sclerotinia sclerotiorum [J]. Eur J Biochem, 1999, 266(1): 252-259. |
| 17 | Gao J, Qian YC, Wang YF, et al. Production of the versatile cellulase for cellulose bioconversion and cellulase inducer synthesis by genetic improvement of Trichoderma reesei [J]. Biotechnol Biofuels, 2017, 10: 272. |
| 18 | Sun JP, Louise Glass N. Identification of the CRE-1 cellulolytic regulon in Neurospora crassa [J]. PLoS One, 2011, 6(9): e25654. |
| 19 | Portnoy T, Margeot A, Linke RT, et al. The CRE1 carbon catabolite repressor of the fungus Trichoderma reesei: a master regulator of carbon assimilation [J]. BMC Genomics, 2011, 12: 269. |
| 20 | Liu DD, Liu Q, Guo WZ, et al. Development of genetic tools in glucoamylase-hyperproducing industrial Aspergillus niger strains [J]. Biology, 2022, 11(10): 1396. |
| 21 | 马东旭, 李忠辉, 张子恒, 等. 高产单宁酶黑曲霉菌株的常温常压等离子体诱变选育及发酵工艺研究 [J/OL]. 食品与发酵工业, 2024, 1-10. . |
| Ma DX, Li ZH, Zhang ZH, et al. Atmospheric room temperature plasma mutagenesis breeding and fermentation optimization of Aspergillus niger with high yield of tannase [J/OL]. Food and Fermentation Industries, 2024. . | |
| 22 | Yin YR, Sang P, Xian WD, et al. Expression and characteristics of two glucose-tolerant GH1 β-glucosidases from Actinomadura amylolytica YIM 77502T for promoting cellulose degradation [J]. Front Microbiol, 2018, 9: 3149. |
| 23 | 赵君. 黑曲霉胞内β-葡萄糖苷酶的鉴定及其在纤维素降解酶系合成调控中的作用 [D]. 郑州: 河南农业大学, 2021. |
| Zhao J. Identification of intracellular β-glucosidase from Aspergillus Niger and its role in synthesis regulation of cellulose degrading enzyme system [D]. Zhengzhou: Henan Agricultural University, 2021. | |
| 24 | Zhong YT, Gu JH, Shang CY, et al. Sustainable succinic acid production from lignocellulosic hydrolysates by engineered strains of Yarrowia lipolytica at low pH [J]. Bioresour Technol, 2024, 408: 131166. |
| 25 | Wang SY, Cheng AH, Liu FH, et al. Catalytic conversion network for lignocellulosic biomass valorization: a panoramic view [J]. Ind Chem Mater, 2023, 1(2): 188-206. |
| 26 | Teugjas H, Väljamäe P. Selecting β-glucosidases to support cellulases in cellulose saccharification [J]. Biotechnol Biofuels, 2013, 6(1): 105. |
| 27 | Okal EJ, Aslam MM, Karanja JK, et al. Mini review: Advances in understanding regulation of cellulase enzyme in white-rot basidiomycetes [J]. Microb Pathog, 2020, 147: 104410. |
| 28 | Ichinose S, Tanaka M, Shintani T, et al. Increased production of biomass-degrading enzymes by double deletion of creA and creB genes involved in carbon catabolite repression in Aspergillus oryzae [J]. J Biosci Bioeng, 2018, 125(2): 141-147. |
| 29 | Li ZH, Yao GS, Wu RM, et al. Synergistic and dose-controlled regulation of cellulase gene expression in Penicillium oxalicum [J]. PLoS Genet, 2015, 11(9): e1005509. |
| 30 | Driouch H, Sommer B, Wittmann C. Morphology engineering of Aspergillus niger for improved enzyme production [J]. Biotechnol Bioeng, 2010, 105(6): 1058-1068. |
| 31 | Zhao QQ, Zhang Z, Liu ZH, et al. A closed-loop strategy for on-site production of saccharolytic enzymes for lignocellulose biorefinery using internal lignocellulosic hydrolysates [J]. Chem Eng J, 2024, 480: 148272. |
| 32 | Delmas S, Pullan ST, Gaddipati S, et al. Uncovering the genome-wide transcriptional responses of the filamentous fungus Aspergillus niger to lignocellulose using RNA sequencing [J]. PLoS Genet, 2012, 8(8): e1002875. |
| [1] | LI Rui, HU Ting, CHEN Shu-wei, WANG Yao, WANG Ji-ping. Positive Regulation of Anthocyanin Biosynthesis by PfMYB80 Transcription Factor in Perilla frutescens [J]. Biotechnology Bulletin, 2025, 41(6): 243-255. |
| [2] | GUO Tao, AI Li-jiao, ZOU Shi-hui, ZHOU Ling, LI Xue-mei. Functional Study of CjRAV1 from Camellia japonica in Regulating Flowering Delay [J]. Biotechnology Bulletin, 2025, 41(6): 208-217. |
| [3] | CHENG Shan, WANG Hui-wei, CHEN Chen, ZHU Ya-jing, LI Chun-xin, BIE Hai, WANG Shu-feng, CHEN Xian-gong, ZHANG Xiang-ge. Cloning of MYB Transcription Factor Gene CeMYB154 and Analysis of Salt Tolerance Function in Cyperus esculentus [J]. Biotechnology Bulletin, 2025, 41(6): 218-228. |
| [4] | LI Xiao-huan, CHEN Xiang-yu, TAO Qi-yu, ZHU Ling, TANG Ming, YAO Yin-an, WANG Li-jun. Effects of PtoMYB61 on Lignin Biosynthesis and Salt Tolerance in Populus tomentosa [J]. Biotechnology Bulletin, 2025, 41(6): 284-296. |
| [5] | LUO Si-fang, ZHANG Zu-ming, XIE Li-fang, GUO Zi-jing, CHEN Zhao-xing, YANG Yue-hua, YAN Xiang, ZHANG Hong-ming. Genome-wide Identification of GATA Gene Family of Jindou Kumquat (Fortunella hindsii) and Their Expression Analysis in Fruit Development [J]. Biotechnology Bulletin, 2025, 41(5): 218-230. |
| [6] | YANG Chun, WANG Xiao-qian, WANG Hong-jun, CHAO Yue-hui. Cloning, Subcellular Localization and Expression Analysis of MtZHD4 Gene from Medicago truncatula [J]. Biotechnology Bulletin, 2025, 41(5): 244-254. |
| [7] | HU Ruo-qun, ZENG Jing-jing, LIANG Wan-feng, CAO Jia-yu, HUANG Xiao-wei, LIANG Xiao-ying, QIU Ming-yue, CHEN Ying. Integrated Transcriptome and Metabolome Analysis to Explore the Carotenoid Synthesis and Metabolism Mechanism in Anoectochilus roxburghii under Different Shading Conditions [J]. Biotechnology Bulletin, 2025, 41(5): 231-243. |
| [8] | QU Shan, ZHAO Yue, LI Ya-hua, ZHENG Gui-ling, XIAN Hong-quan. A Study on the Interaction between Transcriptional Factor and Protein of Tachi2 Chitinase Gene in Trichoderma asperellum [J]. Biotechnology Bulletin, 2025, 41(5): 310-319. |
| [9] | YANG Chao-jie, ZHANG Lan, CHEN Hong, HUANG Juan, SHI Tao-xiong, ZHU Li-wei, CHEN Qing-fu, LI Hong-you, DENG Jiao. Functional Identification of the Transcription Factor Gene FtbHLH3 in Regulating Flavonoid Biosynthesis in Fagopyrum tataricum [J]. Biotechnology Bulletin, 2025, 41(4): 134-144. |
| [10] | WANG Tian-xi, YANG Bing-song, PAN Rong-jun, GAI Wen-xian, LIANG Mei-xia. Identification of the Apple PLATZ Gene Family and Functional Study of the MdPLATZ9 Gene [J]. Biotechnology Bulletin, 2025, 41(4): 176-187. |
| [11] | SONG Shu-yi, JIANG Kai-xiu, LIU Huan-yan, HUANG Ya-cheng, LIU Lin-ya. Identification of the TCP Gene Family in Actinidia chinensis var. Hongyang and Their Expression Analysis in Fruit [J]. Biotechnology Bulletin, 2025, 41(3): 190-201. |
| [12] | WANG Bin, WANG Yu-kun, XIAO Yan-hui. Comparative Transcriptomic Analysis of Clove Basil (Ocimum gratissimum) Leaves in Response to Cadmium Stress [J]. Biotechnology Bulletin, 2025, 41(3): 255-270. |
| [13] | LIU Jie, WANG Fei, TAO Ting, ZHANG Yu-jing, CHEN Hao-ting, ZHANG Rui-xing, SHI Yu, ZHANG Yi. Overexpression of SlWRKY41 Improves the Tolerance of Tomato Seedlings to Drought [J]. Biotechnology Bulletin, 2025, 41(2): 107-118. |
| [14] | LI Yan-wei, YANG Yan-yan, SUN Ya-ling, HUO Yu-meng, WANG Zhen-bao, LIU Bing-jiang. Regulation Mechanism of Plant Hormones Related to Onion Bulb Enlargement and Development Based on Transcriptome Analysis [J]. Biotechnology Bulletin, 2025, 41(2): 187-201. |
| [15] | QIAN Zheng-yi, WU Shao-fang, CAO Shu-yi, SONG Ya-xin, PAN Xin-feng, LI Zhao-wei, FAN Kai. Identification of the NAC Transcription Factors in Nymphaea colorata and Their Expression Analysis [J]. Biotechnology Bulletin, 2025, 41(2): 234-247. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||