








Biotechnology Bulletin ›› 2025, Vol. 41 ›› Issue (8): 276-288.doi: 10.13560/j.cnki.biotech.bull.1985.2025-0116
WEI Yao1( ), ZHANG Jing-jing2, CUI Yun-xiao1, LIU Yu1, LIU Hai-rui1(
), ZHANG Jing-jing2, CUI Yun-xiao1, LIU Yu1, LIU Hai-rui1( )
)
Received:2025-01-28
Online:2025-08-26
Published:2025-08-14
Contact:
LIU Hai-rui
E-mail:1935589234@qq.com;lhybotany@163.com
WEI Yao, ZHANG Jing-jing, CUI Yun-xiao, LIU Yu, LIU Hai-rui. Phylogenetic Analysis of Lonicera Sect. Nintooa Based on Chloroplast Genomes Data[J]. Biotechnology Bulletin, 2025, 41(8): 276-288.
基因功能分类 Category | 基因类别 Gene group | 基因名称 Gene name |
|---|---|---|
| 光合作用有关 Related to photosynthesis | 光系统Ⅰ Photosystem I | psaA, psaB, psaC, psaI, psaJ |
光系统Ⅱ Photosystem Ⅱ | psbA, psbB, psbC, psbD, psbE, psbF, psbH, psbI, psbJ, psbK, psbL, psbM, psbN, psbT, psbZ | |
NADH氧化还原酶基因 NADH dehydrogenase | ndhA*, ndhB*(2), ndhC, ndhD, ndhE, ndhF, ndhG, ndhH, ndhI, ndhJ, ndhK | |
细胞色素b/f复合体 Cytochrome b/f complex | petA, petB*, petD*, petG, petL, petN | |
ATP酶 ATP synthase | atpA, atpB, atpE, atpF*, atpH, atpI | |
二磷酸核酮糖羧化酶大亚基 Rubis CO large subunit | rbcL | |
自我复制基因 Self-replication gene | 核糖体蛋白大亚基 Large subunit of ribosome | rpl14, rpl16*, rpl2*(2), rpl20, rpl22, rpl23, rpl32, rpl33, rpl36 |
核糖体蛋白小亚基 Small subunit of ribosome | rps11, rps12**(2), rps14, rps15, rps16*, rps18, rps19, rps2, rps3, rps4, rps7(2), rps8 | |
RNA聚合酶 RNA polymerase | rpoA, rpoB, rpoC1*, rpoC2 | |
核糖体RNAs Ribosomal RNAs | rrn16(2), rrn23(2), rrn4.5(2), rrn5(2) | |
转运RNAs Transfer RNAs | trnA-UGC*(2), trnC-GCA, trnD-GUC, trnE-UUC, trnF-GAA, trnG-GCC, trnG-UCC*, trnH-GUG*, trnI-CAU(2), trnI-GAU*(2), trnK-UUU*, trnL-CAA(2), trnL-UAA*, trnL-UAG, trnM-CAU, trnN-GUU(2), trnP-UGG, trnQ-UUG, trnR-ACG(2), trnR-UCU, trnS-GCU, trnS-GGA, trnS-UGA, trnT-GGU, trnT-UGU, trnV-GAC(2), trnV-UAC*, trnW-CCA, trnY-GUA, trnfM-CAU | |
其他基因 Other genes | 成熟酶 Maturase | matK |
蛋白酶 Protease | clpP | |
包膜蛋白基因 Envelop membrane protein | cemA | |
C型细胞色素合成基因 C-type cytochrome synthesis gene | ccsA | |
转录起始因子 Translation initiation factor | infA | |
未知基因 Unknown gene | 假定叶绿体阅读框 Hypothetical chloroplast reading frames | ycf1, ycf15(2), ycf2(2), ycf3**, ycf4 |
Table 1 Classification and statistics of chloroplast gene function of Sect. Nintooa
基因功能分类 Category | 基因类别 Gene group | 基因名称 Gene name |
|---|---|---|
| 光合作用有关 Related to photosynthesis | 光系统Ⅰ Photosystem I | psaA, psaB, psaC, psaI, psaJ |
光系统Ⅱ Photosystem Ⅱ | psbA, psbB, psbC, psbD, psbE, psbF, psbH, psbI, psbJ, psbK, psbL, psbM, psbN, psbT, psbZ | |
NADH氧化还原酶基因 NADH dehydrogenase | ndhA*, ndhB*(2), ndhC, ndhD, ndhE, ndhF, ndhG, ndhH, ndhI, ndhJ, ndhK | |
细胞色素b/f复合体 Cytochrome b/f complex | petA, petB*, petD*, petG, petL, petN | |
ATP酶 ATP synthase | atpA, atpB, atpE, atpF*, atpH, atpI | |
二磷酸核酮糖羧化酶大亚基 Rubis CO large subunit | rbcL | |
自我复制基因 Self-replication gene | 核糖体蛋白大亚基 Large subunit of ribosome | rpl14, rpl16*, rpl2*(2), rpl20, rpl22, rpl23, rpl32, rpl33, rpl36 |
核糖体蛋白小亚基 Small subunit of ribosome | rps11, rps12**(2), rps14, rps15, rps16*, rps18, rps19, rps2, rps3, rps4, rps7(2), rps8 | |
RNA聚合酶 RNA polymerase | rpoA, rpoB, rpoC1*, rpoC2 | |
核糖体RNAs Ribosomal RNAs | rrn16(2), rrn23(2), rrn4.5(2), rrn5(2) | |
转运RNAs Transfer RNAs | trnA-UGC*(2), trnC-GCA, trnD-GUC, trnE-UUC, trnF-GAA, trnG-GCC, trnG-UCC*, trnH-GUG*, trnI-CAU(2), trnI-GAU*(2), trnK-UUU*, trnL-CAA(2), trnL-UAA*, trnL-UAG, trnM-CAU, trnN-GUU(2), trnP-UGG, trnQ-UUG, trnR-ACG(2), trnR-UCU, trnS-GCU, trnS-GGA, trnS-UGA, trnT-GGU, trnT-UGU, trnV-GAC(2), trnV-UAC*, trnW-CCA, trnY-GUA, trnfM-CAU | |
其他基因 Other genes | 成熟酶 Maturase | matK |
蛋白酶 Protease | clpP | |
包膜蛋白基因 Envelop membrane protein | cemA | |
C型细胞色素合成基因 C-type cytochrome synthesis gene | ccsA | |
转录起始因子 Translation initiation factor | infA | |
未知基因 Unknown gene | 假定叶绿体阅读框 Hypothetical chloroplast reading frames | ycf1, ycf15(2), ycf2(2), ycf3**, ycf4 |
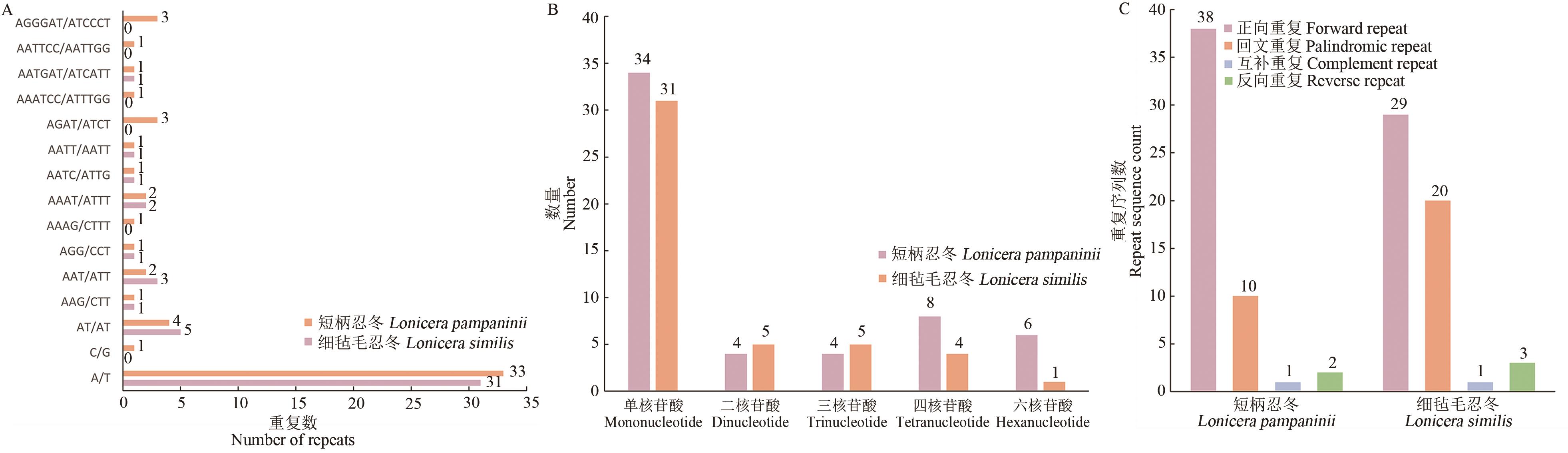
Fig. 6 Simple repetitive sequence analysisA: Number of SSRs in different types. B. Number of nucleotide in different types. C. Number of long repeat sequence
| [1] | 朱颖, 董玉芝, 昝少平, 等. 忍冬科的系统发育核糖体DNA ITS区序列证据 [J]. 干旱区研究, 2007, 24(6): 6840-6844. |
| Zhu Y, Dong YZ, Zan SP, et al. Phylogeny of Caprifoliaceae inferred from the sequence analysis of the nrDNA ITS region [J]. Arid Zone Res, 2007, 24(6): 6840-6844. | |
| [2] | Rehder A. Synopsis of the Genus Lonicera [M]. Saint. Louis: Missouri Botanical Garden, 1903. |
| [3] | 苏金强, 林秀香, 黄阿凤. 福建省忍冬科野生观赏植物资源及其利用 [J]. 福建热作科技, 2005, 30(1): 21-22. |
| Su JQ, Lin XX, Huang AF. Resources and their utilization of the wild ornamental plants of hongeysuckle in Fujian [J]. Fujian Sci Technol Trop Crops, 2005, 30(1): 21-22. | |
| [4] | 方建新. 安徽忍冬科野生植物资源及利用 [J]. 中国农学通报, 2006, 22(5): 412-415. |
| Fang JX. The wilding plants resource of Caprifoliaceae in Anhui Province and their exploitation [J]. Chin Agric Sci Bull, 2006, 22(5): 412-415. | |
| [5] | 王红娟, 何定萍. 重庆市忍冬科野生植物资源及其利用 [J]. 西南园艺, 2006(5): 29-31. |
| Wang HJ, He DP. Wild plant resources of Caprifoliaceae in Chongqing and their utilization [J]. Southwest Hortic, 2006(5): 29-31. | |
| [6] | 刘安成, 毕毅, 尉倩, 等. 中国忍冬属植物育种研究进展 [J]. 北方园艺, 2022(5): 133-139. |
| Liu AC, Bi Y, Yu Q, et al. Research progress on breeding of Lonicera linn.in China [J]. North Hortic, 2022(5): 133-139. | |
| [7] | 安丰云, 金美兰, 玄永男, 等. 蓝靛果忍冬科技生产示范园的建立与研究 [J]. 中国林副特产, 2009(3): 15-18. |
| An FY, Jin ML, Xuan YN, et al. Establishment and studies of technological production demonstration garden of Lonicera caerulea L [J]. For Prod Speciality China, 2009(3): 15-18. | |
| [8] | 李守忠. 黑河市蓝靛果忍冬发展前景初探 [J]. 防护林科技, 2019(3): 83-84. |
| Li SZ. Development prospect of Lonicera edulis in Heihe City [J]. Prot For Sci Technol, 2019(3): 83-84. | |
| [9] | Craves JA. Native birds exploit leaf-mining moth larvae using a new North American host, non-native Lonicera maackii [J]. Écoscience, 2017, 24(3/4): 81-90. |
| [10] | 雷志钧, 周日宝, 童巧珍, 等. 灰毡毛忍冬与正品金银花安全性比较 [J]. 中成药, 2006, 28(5): 759-761. |
| Lei ZJ, Zhou RB, Tong QZ, et al. Comparison of safety between Lonicera macranthoides and genuine honeysuckle [J]. Chin Tradit Pat Med, 2006, 28(5): 759-761. | |
| [11] | 国家药典委员会. 中华人民共和国药典-一部: 2020年版 [M]. 北京: 中国医药科技出版社, 2020. |
| he Pharmacopoeia Committee. People’s republic of China (PRC) pharmacopoeia-part I: 2020 edition [M]. Beijing: China Medical Science Press, 2020. | |
| [12] | 刘婧慧. 金银花与山银花质量评价及安全性对比研究 [D]. 北京: 北京中医药大学, 2014. |
| Liu JH. Comparative study on quality evaluation and safety of Lonicera japonica Flos and Lonicera Flos [D]. Beijing: Beijing University of Chinese Medicine, 2014. | |
| [13] | Theis N, Donoghue MJ, Li JH. Phylogenetics of the caprifolieae and Lonicera (Dipsacales) based on nuclear and chloroplast DNA sequences [J]. Issn, 2008, 33(4): 776-783. |
| [14] | Nakaji M, Tanaka N, Sugawara T. A molecular phylogenetic study of Lonicera L.(Caprifoliaceae) in Japan based on chloroplast DNA sequences[J]. Acta Phytotaxon Geobot, 2015, 66(3): 137-151. |
| [15] | Liu ML, Fan WB, Wang N, et al. Evolutionary analysis of plastid genomes of seven Lonicera L. species: implications for sequence divergence and phylogenetic relationships [J]. Int J Mol Sci, 2018, 19(12): 4039. |
| [16] | Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability [J]. Mol Biol Evol, 2013, 30(4): 772-780. |
| [17] | Sun QH, Morales-Briones DF, Wang HX, et al. Target sequence capture data shed light on the deeper evolutionary relationships of subgenus Chamaecerasus in Lonicera (Caprifoliaceae) [J]. Mol Phylogenet Evol, 2023, 184: 107808. |
| [18] | Yang XL, Sun QH, Morales-Briones DF, et al. New insights into infrageneric relationships of Lonicera (Caprifoliaceae) as revealed by nuclear ribosomal DNA cistron data and plastid phylogenomics [J]. J Syst Evol, 2024, 62(3): 333-357. |
| [19] | Jin JJ, Yu WB, Yang JB, et al. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes [J]. Genome Biol, 2020, 21(1): 241. |
| [20] | Zheng SY, Poczai P, Hyvönen J, et al. Chloroplot: an online program for the versatile plotting of organelle genomes [J]. Front Genet, 2020, 11: 576124. |
| [21] | 李佳, 李清. 美国黄松和蓝粉云杉叶绿体基因组序列测定与分析 [J].植物科学学报, 2022, 40(6): 791-800. |
| Li J, Li Q. Complete chloroplast genome sequence and analysis of Pinus ponderosa P. Lawson & C. Lawson and Picea pungens Engelm [J]. Plant Science Journal, 2022, 40( 6): 791-800. | |
| [22] | Amiryousefi A, Hyvönen J, Poczai P. IRscope: an online program to visualize the junction sites of chloroplast genomes [J]. Bioinformatics, 2018, 34(17): 3030-3031. |
| [23] | Darling ACE, Mau B, Blattner FR, et al. Mauve: multiple alignment of conserved genomic sequence with rearrangements [J]. Genome Res, 2004, 14(7): 1394-1403. |
| [24] | Frazer KA, Pachter L, Poliakov A, et al. VISTA: computational tools for comparative genomics [J]. Nucleic Acids Res, 2004, 32(Web Server issue): W273-W279. |
| [25] | 吴民富, 李莎, 高柔敏, 等. 猫尾草叶绿体基因组结构及其解析 [J]. 福建农业学报, 2023, 38(9): 1094-1102. |
| Wu MF, Li S, Gao RM, et al. Structure of Uraria crinita chloroplast genome [J]. Fujian J Agric Sci, 2023, 38(9): 1094-1102. | |
| [26] | 朱陇强, 朱志煌, 朱雷宇, 等. 匙指虾科线粒体基因组结构与系统进化分析 [J]. 大连海洋大学学报, 2022, 37(3): 428-434. |
| Zhu LQ, Zhu ZH, Zhu LY, et al. Characteristics and phylogenetic analysis of mitogenome in Atyidae [J]. J Dalian Ocean Univ, 2022, 37(3): 428-434. | |
| [27] | 张东, 李文祥, 王桂堂. PhyloSuite: 生物领域分子系统发育分析新平台 [J]. 科学观察, 2023, 18(6): 35-37. |
| Zhang D, Li WX, Wang GT. PhyloSuite: a novel biological platform for molecular phylogenetics studies [J]. Sci Focus, 2023, 18(6): 35-37. | |
| [28] | Xia XM, Yang MQ, Li CL, et al. Spatiotemporal evolution of the global species diversity of Rhododendron [J]. Mol Biol Evol, 2022, 39(1): msab314. |
| [29] | Park YJ, Kang H, Lee HR, et al. The complete chloroplast genome and characteristics analysis of Lonicera harae (Caprifoliaceae) [J]. J Asia Pac Biodivers, 2024, 17(3): 590-594. |
| [30] | 江媛, 杨青淑, 王婧, 等. 毛重楼叶绿体基因组序列特征及其系统发育分析 [J]. 中草药, 2021, 52(13): 4014-4022. |
| Jiang Y, Yang QS, Wang J, et al. Complete chloroplast genome of Paris mairei: characterization and phylogeny [J]. Chin Tradit Herb Drugs, 2021, 52(13): 4014-4022. | |
| [31] | Chen JL, Wang F, Zhao Z, et al. Complete chloroplast genomes and comparative analyses of three Paraphalaenopsis (Aeridinae, Orchidaceae) species [J]. Int J Mol Sci, 2023, 24(13): 11167. |
| [32] | 卢政阳, 于凤扬, 肖月娥, 等. 北陵鸢尾叶绿体基因组及其特征分析 [J]. 草地学报, 2023, 31(6): 1656-1664. |
| Lu ZY, Yu FY, Xiao YE, et al. Complete chloroplast genomes and characteristics analysis of iris typhifolia [J]. Acta Agrestia Sin, 2023, 31(6): 1656-1664. | |
| [33] | 魏亚楠, 龚明贵, 白娜, 等. 梁山慈竹叶绿体基因组密码子偏好性分析 [J]. 浙江农林大学学报, 2024, 41(4): 696-705. |
| Wei YN, Gong MG, Bai N, et al. Analysis of codon preference in chloroplast genome of Dendrocalamus farinosus [J]. J Zhejiang A F Univ, 2024, 41(4): 696-705. | |
| [34] | 李连星, 彭劲谕, 王大玮, 等. 长爪栘[木衣]叶绿体基因组特征系统发育及密码子偏好性分析 [J]. 生物工程学报, 2022, 38(1): 328-342. |
| Li LX, Peng JY, Wang DW, et al. Chloroplast genome phylogeny and codon preference of Docynia longiunguis [J]. Chin J Biotechnol, 2022, 38(1): 328-342. | |
| [35] | 李玉红, 林荣呈. 叶绿素合成调控研究进展 [J]. 生命科学, 2024, 36(9): 1141-1148. |
| Li YH, Lin RC. Advances in the regulation mechanism of chlorophyll synthesis [J]. Chin Bull Life Sci, 2024, 36(9): 1141-1148. | |
| [36] | Li X, Yang JB, Wang H, et al. Plastid NDH pseudogenization and gene loss in a recently derived lineage from the largest hemiparasitic plant genus Pedicularis (Orobanchaceae) [J]. Plant Cell Physiol, 2021, 62(6): 971-984. |
| [37] | 陶爱恩, 尹成龙, 李娅波, 等. 独蒜兰属叶绿体基因组序列及密码子偏好性分析 [J]. 西南林业大学学报: 自然科学, 2025, 45(2): 66-75. |
| Tao AE, Yin CL, Li YB, et al. Analysis of chloroplast genome sequence and codon preference of Pleione [J]. J Southwest For Univ Nat Sci, 2025, 45(2): 66-75. | |
| [38] | 富贵, 刘晶, 李军乔. 密花香薷叶绿体基因组结构及系统进化分析 [J]. 中草药, 2022, 53(6): 1844-1853. |
| Fu G, Liu J, Li JQ. Characterization of chloroplast genome structure and phyletic evolution of Elsholtzia densa [J]. Chin Tradit Herb Drugs, 2022, 53(6): 1844-1853. | |
| [39] | Huo YX, Zhao YK, Xu LW, et al. An integrated strategy for target SSR genotyping with toleration of nucleotide variations in the SSRs and flanking regions [J]. BMC Bioinformatics, 2021, 22(1): 429. |
| [40] | 曾宪法, 刘畅, 杨小英, 等. 大叶百两金和细柄百两金叶绿体全基因组解析及系统发育分析 [J]. 药学学报, 2023, 58(1): 217-228. |
| Zeng XF, Liu C, Yang XY, et al. Chloroplast genome resolution and phylogenetic analysis of Ardisia crispa var. amplifolia and Ardisia crispa var. dielsii [J]. Acta Pharm Sin, 2023, 58(1): 217-228. | |
| [41] | 涂国章, 张显强. DNA条形码技术在石斛分类鉴定中的应用进展 [J]. 食品安全质量检测学报, 2023, 14(2): 154-160. |
| Tu GZ, Zhang XQ. Application progress of DNA barcoding technology in Dendrobium classification and identification [J]. J Food Saf Qual, 2023, 14(2): 154-160. | |
| [42] | 张晶晶, 袁庆, 魏瑶, 等. 忍冬属囊管组植物叶绿体基因组进化分析 [J]. 中草药, 2024, 55(9): 3085-3097. |
| Zhang JJ, Yuan Q, Wei Y, et al. Evolutionary analysis of chloroplast genomes of Sect. Isika (Lonicera) species [J]. Chin Tradit Herb Drugs, 2024, 55(9): 3085-3097. | |
| [43] | Cai HM, Liu X, Wang WQ, et al. Phylogenetic relationships and biogeography of Asia Callicarpa (Lamiaceae), with consideration of a long-distance dispersal across the Pacific Ocean-insights into divergence modes of pantropical flora [J]. Front Plant Sci, 2023, 14: 1133157. |
| [44] | Huang J, Condamine FL, Han MQ, et al. Miocene climatic fluctuations likely explain the species diversification of Lysionotus (Gesneriaceae) in Pan-Himalaya [J]. J Syst Evol, 2025, 63(1): 134-147. |
| [45] | Lai YJ, Wen J, Zhou ZK, et al. Uplift history and biological evolution of the Himalaya (I) [J]. J Syst Evol, 2025, 63(1): 1-4. |
| [46] | Miao YF, Fang XM, Sun JM, et al. A new biologic paleoaltimetry indicating Late Miocene rapid uplift of northern Tibet Plateau [J]. Science, 2022, 378(6624): 1074-1079. |
| [47] | Huang EH, Chen YX, Fang M, et al. Environmental drivers of plant distributions at global and regional scales [J]. Glob Ecol Biogeogr, 2021, 30(3): 697-709. |
| [1] | AN Chang, XU Wen-bo, LU Lin, LI Deng-lin, YAO Yi-xin, LIN Yan-xiang, YANG Cheng-zi, QIN Yuan, ZHENG Ping. Comparative Analysis of Glehnia littoralis from Different Geographic Regions Based on the Characteristics of Chloroplast Genome [J]. Biotechnology Bulletin, 2025, 41(6): 229-242. |
| [2] | LI Bin, SU Xiang-ping, LIU Chang, WANG Yu-bing, ZHANG Yong-hong, ZHOU Chao, XU Qing. Chloroplast Genome Characteristics and Phylogenetic Analysis of Scrophulariaceae [J]. Biotechnology Bulletin, 2025, 41(3): 240-254. |
| [3] | JIANG Hong-yan, CHEN Shi-chun, LIAO Shu-ran, CHEN Ting-xu, WANG Xiao-qing. The Complete Sequences and Phylogenetic Analysis of Mitochondrial Genomes in Eocanthecona concinna and Picromerus lewisi [J]. Biotechnology Bulletin, 2025, 41(1): 312-323. |
| [4] | LIU Dan-dan, WANG Lei-gang, SUN Ming-hui, JIAO Xiao-yu, WU Qiong, WANG Wen-jie. Genome-wide Identification and Expression Pattern Profiling of the Trehalose-6-phosphate Synthase(TPS)Gene Family in Tea Plant(Camellia sinensis) [J]. Biotechnology Bulletin, 2024, 40(8): 152-163. |
| [5] | WU Cui-cui, XIAO Shui-ping. Genome-wide Identification of HD-Zip Gene Family in Gossypium hirsutum L. and Expression Analysis in Response to Abiotic Stress [J]. Biotechnology Bulletin, 2024, 40(2): 130-145. |
| [6] | YANG Yu-qing, TAN Juan, WANG Fang, PENG Shun-li, CHEN Jie, TAN Ming-yan, LYU Mei-yan, ZHOU Fu-yu, LIU Sheng-chuan. Research and Application Progress in Chloroplast Genome of Tea Plant(Camellia sinensis) [J]. Biotechnology Bulletin, 2024, 40(2): 20-30. |
| [7] | CHEN Chao, ZHOU Bo, HE Li, HUO Yan-li, XIAO Fan, LI Chuan-yong, YANG Huan. Research Status and Application of Polysaccharides from Lyophyllum decastes [J]. Biotechnology Bulletin, 2024, 40(11): 14-23. |
| [8] | WANG Zi-xuan, SUI Yu, SHANG Xue-yu, MA Si-jia, WU Tian-xiang, LIU Yang, WANG Qi. Structural Characteristics of Sarcomyxa edulis Polysaccharide and Its Mechanism of Regulating Macrophage Immunomodulatory Activity [J]. Biotechnology Bulletin, 2024, 40(1): 308-321. |
| [9] | YIN Ming-hua, YU Huan-yuan, XIAO Xin-yi, WANG Yu-ting. Chloroplast Genomic Characterization and Phylogenetic Analysis of Colocasia esculenta L. Schoot var. cormosus cv. ‘Hongyayu’ from Jiangxi Yanshan [J]. Biotechnology Bulletin, 2023, 39(6): 233-247. |
| [10] | QU Chun-juan, ZHU Yue, JIANG Chen, QU Ming-jing, WANG Xiang-yu, LI Xiao. Whole Mitochondrial Genome and Phylogeny Analysis of Anomala corpulenta [J]. Biotechnology Bulletin, 2023, 39(2): 263-273. |
| [11] | LIN Xing-yu, SONG Nan. Comparative Mitochondrial Genome and Phylogenetic Analysis of Atteva charopis Turner, 1903 [J]. Biotechnology Bulletin, 2023, 39(12): 300-310. |
| [12] | CHEN Guang-xia, LI Xiu-jie, JIANG Xi-long, SHAN Lei, ZHANG Zhi-chang, LI Bo. Research Progress in Plant Small Signaling Peptides Involved in Abiotic Stress Response [J]. Biotechnology Bulletin, 2023, 39(11): 61-73. |
| [13] | LIU Xiong-wei, LIU Chang, ZENG Xian-fa, YANG Xiao-ying, FENG Ting-ting, ZHAO Jie-hong, ZHOU Ying. Comparative and Phylogenetic Analyses of Complete Chloroplast Genomes in Ardisia crenata [J]. Biotechnology Bulletin, 2023, 39(1): 232-242. |
| [14] | LIU Jing-ju, ZHANG Yu-sen, CHEN Juan, SUN Bing-da, ZHAO Guo-zhu. Research Progress in Modern Taxonomy and Nomenclature of Aspergillus [J]. Biotechnology Bulletin, 2022, 38(7): 109-118. |
| [15] | WANG Chen-chen, ZHANG Fan-li, CHEN Pei-qi, WENG Si-yao, WANG Hui-fang, CUI Xiao-juan. Research Progress in the Structural and Functional Analysis of Mammalian DNA Methyltransferase DNMT1 and DNMT3 [J]. Biotechnology Bulletin, 2022, 38(7): 31-39. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||