








Biotechnology Bulletin ›› 2025, Vol. 41 ›› Issue (9): 115-123.doi: 10.13560/j.cnki.biotech.bull.1985.2025-0346
DONG Xiang-xiang( ), MIAO Bai-ling, XU He-juan, CHEN Juan-juan, LI Liang-jie, GONG Shou-fu, ZHU Qing-song(
), MIAO Bai-ling, XU He-juan, CHEN Juan-juan, LI Liang-jie, GONG Shou-fu, ZHU Qing-song( )
)
Received:2025-04-02
Online:2025-09-26
Published:2025-09-24
Contact:
ZHU Qing-song
E-mail:dong_xiangx@163.com;zqs43@126.com
DONG Xiang-xiang, MIAO Bai-ling, XU He-juan, CHEN Juan-juan, LI Liang-jie, GONG Shou-fu, ZHU Qing-song. Bioinformatics Analysis and Flowering Regulation Function of FveBBX20 Gene in Woodland Strawberry[J]. Biotechnology Bulletin, 2025, 41(9): 115-123.
基因名称 Name of gene | 正向序列 Positive sequence (5′-3′) | 反向序列 Reverse sequence (5′-3′) |
|---|---|---|
| FveBBX20 | ATGAAGATCCGGTGCAATTTTT | TATGTAAGGTGCACTTTTAGCCTCA |
| AtACT2 | AAGCTGGGGTTTTATGAATGG | TTGTCACACACAAGTGCATCAT |
| Fve26S | TAACCGCATCAGGTCTCCAA | CTCGAGCAGTTCTCCGACAG |
| AtCO | ATTCTGCAAACCCACTTGCT | CCTCCTTGGCATCCTTATCA |
| AtFT | CTGGAACAACCTTTGGCAAT | AGCCACTCTCCCTCTGACAA |
| AtFUL | GGAGAAGAAAACGGGTCAGC | ACTCGTTCGTAGTGGTAGGAC |
| AtSOC1 | AATTCGCCAGCTCCAATATG | CCTCGATTGAGCATGTTCCTA |
Table 1 Primer Information
基因名称 Name of gene | 正向序列 Positive sequence (5′-3′) | 反向序列 Reverse sequence (5′-3′) |
|---|---|---|
| FveBBX20 | ATGAAGATCCGGTGCAATTTTT | TATGTAAGGTGCACTTTTAGCCTCA |
| AtACT2 | AAGCTGGGGTTTTATGAATGG | TTGTCACACACAAGTGCATCAT |
| Fve26S | TAACCGCATCAGGTCTCCAA | CTCGAGCAGTTCTCCGACAG |
| AtCO | ATTCTGCAAACCCACTTGCT | CCTCCTTGGCATCCTTATCA |
| AtFT | CTGGAACAACCTTTGGCAAT | AGCCACTCTCCCTCTGACAA |
| AtFUL | GGAGAAGAAAACGGGTCAGC | ACTCGTTCGTAGTGGTAGGAC |
| AtSOC1 | AATTCGCCAGCTCCAATATG | CCTCGATTGAGCATGTTCCTA |

Fig. 2 Bioinformatics analysis of FveBBX20 proteinA: CDS and amino acid sequence of FveBBX20 gene. B: Analysis of secondary structure from FveBBX20 protein. C: Analysis of tertiary structure from FveBBX20 protein. E: Structural domain of FveBBX20 protein. F: Multiple sequence comparison of BBX20 proteins
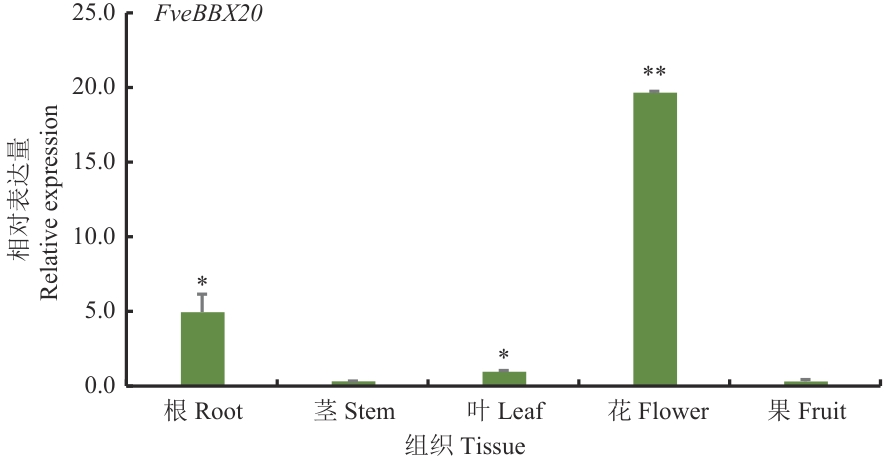
Fig. 4 Relative expressions of FveBBX20 in different tissues* indicates statistically significant differences at P<0.05 level, and ** indicates statistically significant differences at P<0.01 level. The same below

Fig. 5 Identification and flowering function analysis of A. thaliana over-expressing FveBBX20 geneA: PCR identification of FveBBX20 in transgenic A. thaliana, M. DL5000 marker, 1-3. overexpression line, 4. wild typeCol-0. B: Expression analysis of FveBBX20 gene in transgenic A. thaliana. C: The flowering phenotype of transgenic lines. D: Statistics of bolting time of transgenic lines, different lowercase letters indicating significant differences between different strains (P<0.05). E-H: Expression level analysis of flowering-related genes
| [1] | Gangappa SN, Botto JF. The BBX family of plant transcription factors [J]. Trends Plant Sci, 2014, 19(7): 460-470. |
| [2] | 刘嘉, 韩峰, 王雪源, 等. B-box家族蛋白在植物光信号转导中的作用机制进展 [J]. 植物生理学报, 2022, 58(6): 1023-1036. |
| Liu J, Han F, Wang XY, et al. Recent advances on BBX protein family mediated light signal transduction in plants [J]. Plant Physiol J, 2022, 58(6): 1023-1036. | |
| [3] | Khanna R, Kronmiller B, Maszle DR, et al. The Arabidopsis B-box zinc finger family [J]. Plant Cell, 2009, 21(11): 3416-3420. |
| [4] | Huang JY, Zhao XB, Weng XY, et al. The rice B-box zinc finger gene family: genomic identification, characterization, expression profiling and diurnal analysis [J]. PLoS One, 2012, 7(10): e48242. |
| [5] | Xu XH, Li WL, Yang SK, et al. Identification, evolution, expression and protein interaction analysis of genes encoding B-box zinc-finger proteins in maize [J]. J Integr Agric, 2023, 22(2): 371-388. |
| [6] | Chen SH, Jiang WQ, Yin JL, et al. Genome-wide mining of wheat B-BOX zinc finger (BBX) gene family provides new insights into light stress responses [J]. Crop Pasture Sci, 2021, 72(1): 17. |
| [7] | Feng Z, Li MY, Li Y, et al. Comprehensive identification and expression analysis of B-Box genes in cotton [J]. BMC Genomics, 2021, 22(1): 439. |
| [8] | Fan CM, Hu RB, Zhang XM, et al. Conserved CO-FT regulons contribute to the photoperiod flowering control in soybean [J]. BMC Plant Biol, 2014, 14: 9. |
| [9] | Bu X, Wang XJ, Yan JR, et al. Genome-wide characterization of B-box gene family and its roles in responses to light quality and cold stress in tomato [J]. Front Plant Sci, 2021, 12: 698525. |
| [10] | Wang YY, Zhai ZF, Sun YT, et al. Genome-wide identification of the B- BOX genes that respond to multiple ripening related signals in sweet cherry fruit [J]. Int J Mol Sci, 2021, 22(4): 1622. |
| [11] | Cao J, Yuan JL, Zhang YL, et al. Multi-layered roles of BBX proteins in plant growth and development [J]. Stress Biol, 2023, 3(1): 1. |
| [12] | Song ZQ, Bian YT, Liu JJ, et al. B-box proteins: Pivotal players in light-mediated development in plants [J]. J Integr Plant Biol, 2020, 62(9): 1293-1309. |
| [13] | Tripathi P, Carvallo M, Hamilton EE, et al. Arabidopsis B-BOX32 interacts with CONSTANS-LIKE3 to regulate flowering [J]. Proc Natl Acad Sci USA, 2017, 114(1): 172-177. |
| [14] | Cheng H, Yu Y, Zhai YW, et al. An ethylene-responsive transcription factor and a B-box protein coordinate vegetative growth and photoperiodic flowering in Chrysanthemum [J]. Plant Cell Environ, 2023, 46(2): 440-450. |
| [15] | Wang Q, Wang LJ, Cheng H, et al. Two B-box proteins orchestrate vegetative and reproductive growth in summer Chrysanthemum [J]. Plant Cell Environ, 2024, 47(8): 2923-2935. |
| [16] | Ping Q, Cheng PL, Huang F, et al. The heterologous expression in Arabidopsis thaliana of a Chrysanthemum gene encoding the BBX family transcription factor CmBBX13 delays flowering [J]. Plant Physiol Biochem, 2019, 144: 480-487. |
| [17] | Yang YJ, Ma C, Xu YJ, et al. A zinc finger protein regulates flowering time and abiotic stress tolerance in Chrysanthemum by modulating gibberellin biosynthesis[J]. Plant Cell, 2014, 26(5): 2038-2054. |
| [18] | Tan JJ, Jin MN, Wang JC, et al. OsCOL10, a CONSTANS-like gene, functions as a flowering time repressor downstream of Ghd7 in rice [J]. Plant Cell Physiol, 2016, 57(4): 798-812. |
| [19] | Wu WX, Zheng XM, Chen DB, et al. OsCOL16, encoding a CONSTANS-like protein, represses flowering by up-regulating Ghd7 expression in rice [J]. Plant Sci, 2017, 260: 60-69. |
| [20] | Wu WX, Zhang YX, Zhang M, et al. The rice CONSTANS-like protein OsCOL15 suppresses flowering by promoting Ghd7 and repressing RID1 [J]. Biochem Biophys Res Commun, 2018, 495(1): 1349-1355. |
| [21] | 邵妍丽, 卢贝, 贾思振, 等. 草莓FaWRKY70基因克隆与表达模式分析 [J]. 西北植物学报, 2024, 44(7): 1105-1112. |
| Shao YL. Lu B, Jia SZ,et al. Cloning and expression analysis of the FaWRKY70 gene in strawberry [J]. Acta Botanica Boreali-Occidentalia Sinica, 2024, 44(7): 1105-1112. | |
| [22] | 张杨, 马宗桓, 毛娟, 等. 5种叶面肥对大棚草莓光合特性及品质的影响 [J]. 西北植物学报, 2024, 44(4): 562-571. |
| Zhang Y, Ma ZH, Mao J, et al. Effects of five foliar fertilizers on photosynthetic characteristics and fruit quality of Fragaria ananassa in greenhouse [J]. Acta Botanica Boreali-Occidentalia Sinica, 2024, 44(4): 562-571. | |
| [23] | 叶云天, 娄可飞, 王熙然, 等. 森林草莓B-box基因家族全基因组序列鉴定与表达分析 [J] .分子植物育种, 2016, 14(4): 827-834. |
| Ye YT, Lou KF, Wang XR, et al. Genome-wide identification and expression investigation of the B-box gene family in Fragaria vesca [J]. Molecular Plant Breeding, 2016, 14(4): 827-834. | |
| [24] | Ye YT, Liu YQ, Li XL, et al. An evolutionary analysis of B-box transcription factors in strawberry reveals the role of FaBBx28c1 in the regulation of flowering time [J]. Int J Mol Sci, 2021, 22(21): 11766. |
| [25] | 张奇瑞, 刘小林, 徐胜光, 等. 外源茉莉酸甲酯对连作草莓土壤养分和酶活性的影响 [J]. 西南农业学报, 2022, 35(8): 1764-1769. |
| Zhang QR, Liu XL, Xu SG, et al. Effects of exogenous methyl jasmonate on soil nutrients and enzyme activities of continuous cropping strawberry [J]. Southwest China Journal of Agricultural Sciences, 2022, 35(8): 1764-1769. | |
| [26] | He WZ, Liu H, Wu Z, et al. The AaBBX21-AaHY5 module mediates light-regulated artemisinin biosynthesis in Artemisia annua L [J]. J Integr Plant Biol, 2024, 66(8): 1735-1751. |
| [27] | 邓巧灵, 华玮, 王华素, 等. 辣椒CcBBX20基因克隆和表达分析 [J]. 辣椒杂志, 2024, 22(1): 1-8. |
| Deng QL, Hua W, Wang HS, et al. Cloning and expression analysis of gene CcBBX20 in pepper [J]. J China Capsicum, 2024, 22(1): 1-8. | |
| [28] | 周昕越, 蒋庆雪, 贾会丽, 等. 紫花苜蓿MsBBX20基因克隆及耐盐功能分析 [J]. 草业学报, 2024, 33(10): 55-73. |
| Zhou XY, Jiang QX, Jia HL, et al. Cloning and salt-tolerance functional analysis of alfalfa MsBBX20 gene [J]. Acta Prataculturae Sinica, 2024, 33(10): 55-73. | |
| [29] | 陈新娜, 张利英, 喻锌, 等. 甘菊ClCOL16基因的克隆与功能初步研究 [J]. 西北植物学报, 2023, 43(3): 366-373. |
| Chen XN, Zhang LY, Yu X, et al. Cloning and functional analysis of ClCOL16 from Chrysanthemum lavandulifolium [J]. Acta Bot Boreali Occidentalia Sin, 2023, 43(3): 366-373. | |
| [30] | Kurokura T, Samad S, Koskela E, et al. Fragaria vesca CONSTANS controls photoperiodic flowering and vegetative development [J]. J Exp Bot, 2017, 68(17): 4839-4850. |
| [31] | Mouhu K, Kurokura T, Koskela EA, et al. The Fragaria vesca homolog of suppressor of overexpression of constans1 represses flowering and promotes vegetative growth [J]. Plant Cell, 2013, 25(9): 3296-3310. |
| [32] | Wang CQ, Sarmast MK, Jiang JS, et al. The transcriptional regulator BBX19 promotes hypocotyl growth by facilitating COP1-mediated EARLY FLOWERING3 degradation in Arabidopsis [J]. Plant Cell, 2015, 27(4): 1128-1139. |
| [33] | 黄晓. 荔枝BBX基因响应UV-B和低温调控开花的机理研究 [D]. 广州: 华南农业大学, 2024. |
| Huang X. BBX-mediated regulation of litchi flowering in response to UV-B and low temperature [D]. Guangzhou: South China Agricultural University, 2024. |
| [1] | LI Kai-jie, WU Yao, LI Dan-dan. Cloning of Gene CtbHLH128 in Safflower and Response Function Regulating Drought Stress [J]. Biotechnology Bulletin, 2025, 41(8): 234-241. |
| [2] | LAI Shi-yu, LIANG Qiao-lan, WEI Lie-xin, NIU Er-bo, CHEN Ying-e, ZHOU Xin, YANG Si-zheng, WANG Bo. The Role of NbJAZ3 in the Infection of Nicotiana benthamiana by Alfalfa Mosaic Virus [J]. Biotechnology Bulletin, 2025, 41(8): 186-196. |
| [3] | KANG Qin, WANG Xia, SHEN Ming-yang, XU Jing-tian, CHEN Shi-lan, LIAO Ping-yang, XU Wen-zhi, WU Wei, XU Dong-bei. Cloning and Expression Analysis of the UV-B Receptor Gene McUVR8 in Mentha canadensis L. [J]. Biotechnology Bulletin, 2025, 41(8): 255-266. |
| [4] | LA Gui-xiao, ZHAO Yu-long, DAI Dan-dan, YU Yong-liang, GUO Hong-xia, SHI Gui-xia, JIA Hui, YANG Tie-gang. Identification of Plasma Membrane H+-ATPase Gene Family in Safflower and Expression Analysis in Response to Low Nitrogen and Low Phosphorus Stress [J]. Biotechnology Bulletin, 2025, 41(8): 220-233. |
| [5] | WEI Yu-jia, LI Yan, KANG Yu-han, GONG Xiao-nan, DU Min, TU Lan, SHI Peng, YU Zi-han, SUN Yan, ZHANG Kun. Cloning and Expression Analysis of the CrMYB4 Gene in Carex rigescens [J]. Biotechnology Bulletin, 2025, 41(7): 248-260. |
| [6] | LI Xin-ni, LI Jun-yi, MA Xue-hua, HE Wei, LI Jia-li, YU Jia, CAO Xiao-ning, QIAO Zhi-jun, LIU Si-chen. Identification of the PMEI Gene Family of Pectin Methylesterase Inhibitor in Foxtail Millet and Analysis of Its Response to Abiotic Stress [J]. Biotechnology Bulletin, 2025, 41(7): 150-163. |
| [7] | CHENG Shan, WANG Hui-wei, CHEN Chen, ZHU Ya-jing, LI Chun-xin, BIE Hai, WANG Shu-feng, CHEN Xian-gong, ZHANG Xiang-ge. Cloning of MYB Transcription Factor Gene CeMYB154 and Analysis of Salt Tolerance Function in Cyperus esculentus [J]. Biotechnology Bulletin, 2025, 41(6): 218-228. |
| [8] | PEI Jing-qi, ZHAO Meng-ran, HUANG Chen-yang, WU Xiang-li. Discovery and Verification of a Functional Gene Influencing the Growth and Development of Pleurotus ostreatus [J]. Biotechnology Bulletin, 2025, 41(6): 327-334. |
| [9] | XU Hui-zhen, SHANTWANA Ghimire, RAJU Kharel, YUE Yun, SI Huai-jun, TANG Xun. Analysis of the Potato SUMO E3 Ligase Gene Family and Cloning and Expression Pattern of StSIZ1 [J]. Biotechnology Bulletin, 2025, 41(6): 119-129. |
| [10] | LIU Xin, WANG Jia-wen, LI Jin-wei, MOU Ce, YANG Pan-pan, MING Jun, XU Lei-feng. Cloning and Expression Analysis of Three LdBBXs in Lilium davidii var. willmottiae [J]. Biotechnology Bulletin, 2025, 41(5): 186-196. |
| [11] | LI Zhi-qiang, LUO Zheng-qian, XU Lin-li, ZHOU Guo-hui, QU Si-yu, LIU En-liang, XU Dong-ting. Identification of the Soybean R2R3-MYB Gene Family Based on the T2T Genome and Their Expression Analysis under Drought and Salt Stress [J]. Biotechnology Bulletin, 2025, 41(5): 141-152. |
| [12] | ZHAO Jing, GUO Qian, LI Rui-qi, LEI Ying-yang, YUE Ai-qin, ZHAO Jin-zhong, YIN Cong-cong, DU Wei-jun, NIU Jing-ping. Sequence Analysis and Induced Expression Analysis of GmGST Gene Cluster Genes in Soybean [J]. Biotechnology Bulletin, 2025, 41(5): 129-140. |
| [13] | YANG Chun, WANG Xiao-qian, WANG Hong-jun, CHAO Yue-hui. Cloning, Subcellular Localization and Expression Analysis of MtZHD4 Gene from Medicago truncatula [J]. Biotechnology Bulletin, 2025, 41(5): 244-254. |
| [14] | BAN Qiu-yan, ZHAO Xin-yue, CHI Wen-jing, LI Jun-sheng, WANG Qiong, XIA Yao, LIANG Li-yun, HE Wei, LI Ye-yun, ZHAO Guang-shan. Cloning of Phytochrome Interaction Factor CsPIF3a and Its Response to Light and Temperature Stress in Camellia sinensis [J]. Biotechnology Bulletin, 2025, 41(4): 256-265. |
| [15] | LU Yong-jie, XIA Hai-qian, LI Yong-ling, ZHANG Wen-jian, YU Jing, ZHAO Hui-na, WANG Bing, XU Ben-bo, LEI Bo. Cloning and Expression Analysis of AP2/ERF Transcription Factor NtESR2 in Nicotiana tabacum [J]. Biotechnology Bulletin, 2025, 41(4): 266-277. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||